Seasons Greetings
All the best for Christmas and every success in the New Year,
Cheers
Chris

All monies saved by using an electronic card will be donated to multiple sclerosis research
SureChEMBL
I just saw a note that the ChEMBL group that they are taking over the running of the SureChem system from Digital Science. This means that ChEMBL will be collating the chemical patent literature.
More details from the announcement
For those of you that are already SureChem users you will be familiar with the functionality and how it works; but for those that weren't SureChEMBL takes feeds of full text patents, identifies chemical objects from either the in-line text or from images and adds 2-D chemical structures. This is then loaded into a database and is searchable by chemical structure, so you can do substructure, similarity searching and so forth - all the good things you'd expect from a chemical database. This chemical search functionality is unavailable from the public, published patent documents, and is really essential for anyone seriously using the patent literature. Oh, and the system does this live, so as patents are published, they are processed and added to the system - the delay between publication and structures being available in SureChEMBL is about a day when converted from text, and a few days when converted from image sources
Bioisosteres Updated
I’ve just updated the aromatic bioisosteres page to include the bicyclo[1.1.1]pentane replacement for phenyl described in a recent publication DOI.
Published Fragment Hits
I’ve continued to collect details of fragment based screening hits that have been reported in the literature. There are now over 600 hits reported for 113 different targets culled from over 160 publications. I’ll update the calculated properties for those compounds in due course. I was interested in seeing if the physicochemical profiles are different depending on the type of target, however as the plot below shows, the majority of those hits have been identified against enzyme targets so I think I’ll need more data before any meaningful conclusions can be made.
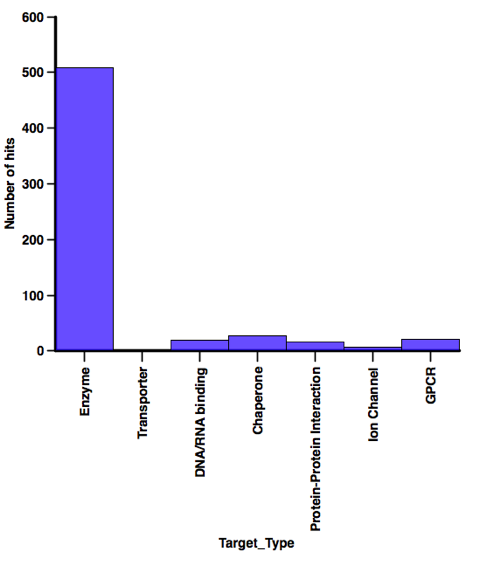
In contrast when looking at the screening technology used a variety of technologies have afforded a substantial number of hits, when I’ve abstracted the latest batch of papers I’ll have a look at the profiles of the compounds identified using each technology.
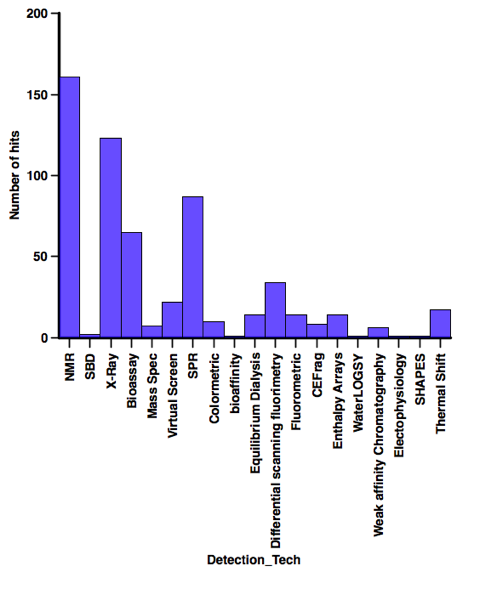
Finding the data is getting more of a challenge, it seems as fragment screening becomes more mainstream it is often not mentioned in the title or abstract. So if you have recently published a relevant paper if you could send me the reference or even a pdf I’d be very grateful.
Drug Discovery Resources Update
I’m in the process of updating the drug discovery resources pages.
I’ve added a couple more examples to the bioisosteres pages and revamped the Computational Chemistry Tools page.
I’ve also updated the Journal RSS feeds and included the feed for Chemical Biology and Drug Design.
Seeding Drug Discovery
The closing date for the next round of the Wellcome Trust Seeding Drug Discovery initiative is November 8 2013.
Funding to facilitate early-stage small-molecule drug discovery. The awards help applicants with a potential drug target or new chemistry embark on a programme of compound discovery and/or take later stage projects towards clinical trials. The aim of Seeding Drug Discovery is to develop drug-like, small molecules that will be the springboard for further research and development by the biotechnology and pharmaceutical industry in areas of unmet medical need.A two-point entry system has been introduced to enable projects at an earlier stage in development to be competitive for funding as well as to progress later-stage projects further towards clinical trials.
Plasma Protein Binding
I’ve just updated the section on distribution and plasma protein binding in the Drug Discovery Resources.
Nitro bioisosteres.
At the 17th RSC-SCI Medicinal Chemistry Conference in Cambridge Alexander Pasternak (Merck) gave an excellent talk on their work to identify a potent and selective ROM-K inhibitor as novel diuretics. The ROM-K potassium channel is a member of the inward rectifier family of potassium channels expressed in two regions of the kidney: thick ascending loop of Henle and cortical collecting duct DOI, ROMK participates in potassium recycling across the luminal membrane which is critical for the function of the Na+/K+/2C1" co-transporter, the rate- determining step for salt reuptake in this part of the nephron. At the cortical collecting duct , ROMK provides a pathway for potassium secretion that is tightly coupled to sodium uptake through the amiloride sensitive sodium channel. This makes ROM-K an attractive potassium sparing diuretic target.
To cut a long story short Merck ran a HTS campaign (actually I think they ran two) and the only hit is shown below.
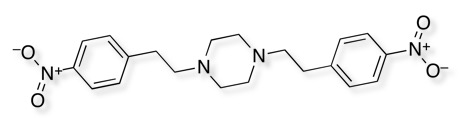
As I am sure all medicinal chemists are aware nitro groups, in particular aromatic nitro groups are well known to be reduced in vivo yielding hydroxylamines and nitrosoamines that are highly reactive species and are known carcinogens. So whilst one nitro in the hit is bad imagine how it feels to have two!
The Merck group however followed this lead up and managed to identify several bioisosteric replacements for the nitro group,
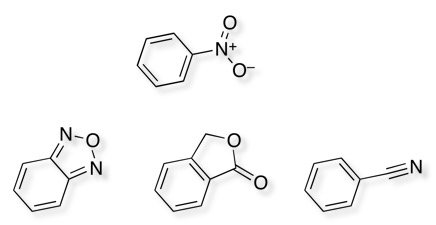
Interestingly there have been two other reported hits for the same target, and these also include nitrobenzenes.
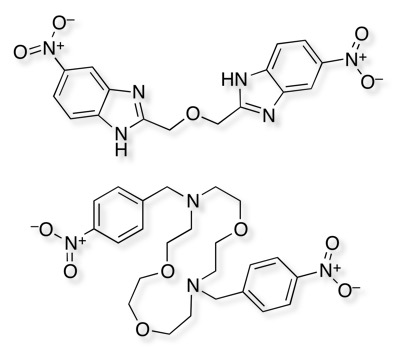
These structures underline the importance of the arylnitro group but also raise a couple of interesting questions, whether nitro compounds should be removed from screening collections? In addition, given the structure of ion channels is often a parallel array of identical proteins forming a pore through the membrane perhaps we should try to populate screening collections with palindromic structures that might bind linking two chains?
I’ll add these to the bioisosteres section at the weekend.
Shape distribution for fragment collections
I’ve now updated the physicochemical property profiles of all the fragment collections I have access to, including the categorisation into rod-, disc- or sphere-like shapes I described last week.
I thought it might be interesting to generate a plot of all 170,000 fragments to look at the distribution. I actually viewed the results in Vortex as shown below. This tool makes it easy to colour by “shape” and also allows me to highlight a few structures that appear at the extremism of the plot.
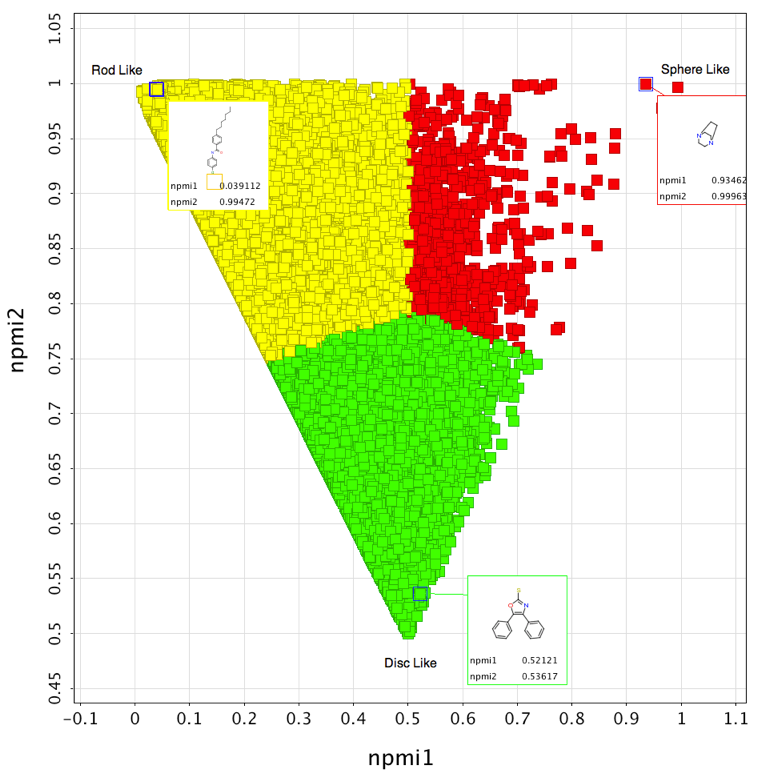
3D Shape distributions of Compound Collections
I recently updated the fragment collections page this included updating the physicochemical property profiles adding npmi (Normalized ratio of principle moments of inertia) as described by Sauer WH, Schwarz MK (2003) Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J Chem Inf Comput Sci 43:987–10030. DOI As the image below shows this gives a view of the shape of the molecules as to whether they are rod, disk or sphere like.
Whilst this works very well for individual compounds or small libraries the plot becomes a blur of overlapping points for larger collections and it is not really possible to compare collections. Whilst it may be possible to generate a single number as the “average” of each collection I’m not sure how useful it would be. So with help from Matt I decided to divide the plot into three sections as shown below.
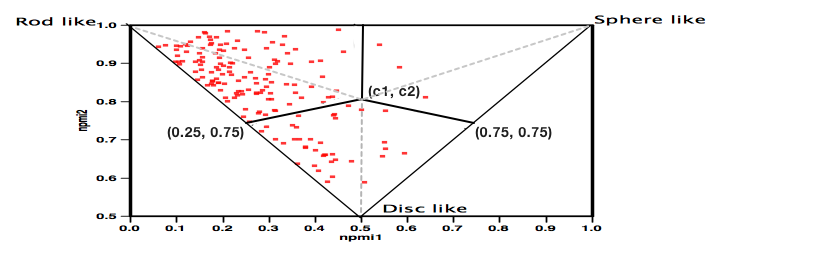
The centre point (c1, c2) was calculated using ( 0.5, (2sqrt(0.5) + 0.5)/(2sqrt(0.5) + 1) ) which is about (0.5, 0.793).
Each of the points in the plot was then assigned to a category using:
If a point is below both lines then: (0.5 - 0.25) * (npmi2 - 0.75) - (0.793 - 0.75) * (npmi1 - 0.25) < 0 and (0.5 - 0.75) * (npmi2 - 0.75) - (0.793 - 0.75) * (npmi1 - 0.75) > 0 then it is disc-like.
If not, then it is rod-like if npmi1 < 0.5 and sphere-like if npmi1 > 0.5.
The result for a 35,000 compound collection are shown below, with the points colour-coded by the assigned category.
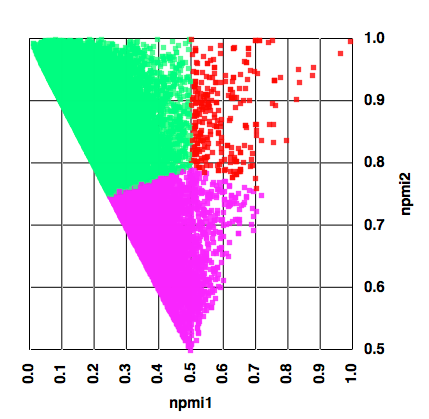
We can then create a categorical plot as shown below.
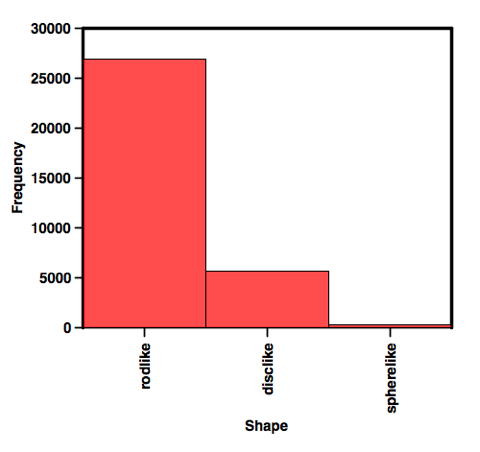
I plan to update all the physicochemical profiles of all the fragment collections next week.
3D Fragment Consortium
I just added the dataset from the 3D Fragment Library Consortium.
The 3D Fragment Consortium brings together UK-based not-for-profit drug discovery institutes and academic groups, working in partnership to build a collection of chemically diverse molecules with a particular focus on fragments that incorporate 3D structure. The consortium is looking to collaborate with other research groups to expand the collection and make it available for screening against new biological targets to help kick-start hit discovery programmes and provide a foundation for a vibrant pre-competitive drug discovery network across the UK. The 3D Fragment Consortium has identified a foundation library of 170 fragments to commence their screening activities.
I’ve added this fragment library profile to the 22 other collections previously calculated:
Comparison of all collections, and calculated physiochemical profiles.
It is obviously early days yet but it will be interesting to see how this develops.
Protein-Protein Interactions
I’ve just added a section on Protein-Protein interactions.
These seem to be an increasingly popular target class and I’ hope to expand on it as we find out more.
How are fragments optimised?
A recent paper J. Med. Chem. 2013, 56, 2478−2486 DOI looked at the different ways that the initial fragments were subsequently optimised, looking at a variety of physicochemical properties and ligand efficiency indices.
I’ve updated the known fragments page to include some of their conclusions.
Open Source Drug Discovery
If you have ever wondered how Open Source Drug Discovery might work there is a very nice example on the Intermolecular blog here.
Well worth a read.
And now for something completely different
In the last few updates I’ve concentrated on Fragment-Based-Screening, where by screening a small set of low molecular weight fragments it is possible to obtain hits that can then be expanded. The latest update highlights the use of encoded libraries, here vast libraries (>>1M compounds) can be screened the hits isolated and identified by virtue of a DNA tag.
From the early “proof-of-principle” peptide libraries this area has now extended into a variety of interesting chemistries.
Biological and MedChem Meeting
I just got details of an interesting meeting in Cambridge, UK later this year.
Known Fragment Hits
I’ve just updated the section describing fragment hits reported in the literature.The dataset now has >500 entries culled from 150 publications directed at nearly 100 different molecular targets using 18 different detection technologies and might be expected to give some insight into the type of compounds that appear as hits.
I’ve calculated a number of physicochemical descriptors and identified fragments that have appeared as hits against multiple targets.
Fragment Pages updated
I’ve just updated the section on fragment libraries, I’ve added a couple of new vendors and updated the existing vendors, there are now over 160,000 fragments available from commercial suppliers. I’ve recalculated the identity and similarity matrix. I’ve also updated the physicochemical property profiles and added npmi (Normalized ratio of principle moments of inertia) as described by Sauer WH, Schwarz MK (2003) Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J Chem Inf Comput Sci 43:987–10030. DOI As the image below shows this gives a view of the shape of the molecules as to whether they are rod, disk or sphere like, it is included with all the other calculated properties.

I notice that a couple of the vendor with very large fragment collections now sell relatively small subsets, underlining the fact that a library of 2000 fragments is usually sufficient as a screening set. Access to the larger fragment space is only really needed when you come to explore the hits.
Separation of PK and PD
I’ve just added a section on compounds with extended off rates from the protein target.
For indications for which require an extended pharmacological profile a compound with a long binding half-life can have a duration of action which extends beyond the presence of drug levels in plasma needed for biological activity. In particular it may be possible to extend duration of action at the intended target whilst limiting activity at off-target proteins. Whilst this could be achieved by irreversible covalent binding there is now a growing body of evidence that many small molecules can display slow off rate kinetics
Learning from our mistakes: The ‘unknown knowns’ in fragment screening
Whilst fragment-based screening has been around for a while there are still groups that are new to the area. This invaluable paper provides an insight into the pit-falls that await the unwary scientist. Absolutely essential reading
In the past 15 years, fragment-based lead discovery (FBLD) has been adopted widely throughout academia and industry. The approach entails discovering very small molecular fragments and growing, merging, or linking them to produce drug leads. Because the affinities of the initial fragments are often low, detection methods are pushed to their limits, leading to a variety of artifacts, false positives, and false negatives that too often go unrecognized. This Digest discusses some of these problems and offers suggestions to avoid them. Although the primary focus is on FBLD, many of the lessons also apply to more established approaches such as high-throughput screening.
Learning from our mistakes: The ‘unknown knowns’ in fragment screening DOI
Published Fragment hits
Whilst there are a variety of techniques to measure the properties or diversity of fragment libraries it is interesting to look at the profiles of compounds that actually appear as hits in fragment-based screening campaigns. I’ve been compiling a database of compounds that have been reported as hits in the literature, this database now has >500 entries culled from 150 publications directed at nearly 100 different molecular targets using 18 different detection technologies and might be expected to give some insight into the type of compounds that appear as hits.
Suggested Books
I’ve just updated the list of suggested books.
Included books on bioisosteres and fragment-based screening.
17th RSC/SCI Medicinal Chemistry Symposium
The registrations are coming in for the forthcoming 17th RSC/SCI Medicinal Chemistry Symposium to be held in Cambridge UK (8-11 Sept 2013). Book early to avoid disappointment.

Full details of the scientific programme are available here together with the registration form.
Fragment Collections
I’ve just updated the page containing the profiles of commercial fragment collections.
Fragment library design: the evolution of fragment-based lead discovery
Latest Publication
Fragment library design: the evolution of fragment-based lead discovery
By Dr E. Zartler, Dr C. Swain & S. Pearce
Drug Discovery World Winter 2012/13
With the growing need to streamline the drug discovery process, screening against fragment libraries rather than drug-like molecules has become increasingly adopted as an integral part of many drug discovery programmes. However, success depends on the quality of the fragment library, and many factors dictate quality.
European Lead Factory
I’m delighted to see the announcement about the European Lead Factory, hopefully this will be a huge asset for Drug Discovery. http://www.nature.com/news/europe-bets-on-drug-discovery-1.12372
Two sites shuttered by the pharmaceutical giant Merck, one in Scotland and one in the Netherlands, will soon be humming again with the work of drug discovery. But the hum will not be business as usual. It will be the sound of a public–private consortium placing a high-stakes wager: a nearly €200-million (US$271-million) bet that it can boost a languishing pharmaceutical sector by fusing academic innovation with industrial-scale screening, using robots to test chemicals for biological activity….Any academic group or company can also propose assays to test molecules in the library for biological activity. Lead-factory scientists will run these assays free of charge and confirm any promising results, working mainly in laboratory space closed by Merck in 2011 at Oss in the Netherlands. Follow-up work will be done at the University of Dundee in Scotland. Results will be provided confidentially to the groups that proposed the assays so that they can pursue further work and publications.
17th RSC/SCI Medicinal Chemistry Symposium
The latest details of the forthcoming 17th RSC/SCI Medicinal Chemistry Symposium to be held in Cambridge UK (8-11 Sept 2013) are now available.

Full details of the scientific programme are available here together with the registration form.
STEM Team East
STEM (Science, Technology, Engineering and Mathematics) Team East is an education charity of 27 years standing. Their aim is to promote STEM education to students.
They provide bespoke consultation on STEM enrichment to each secondary school; teacher CPD in digital electronics and CAD; teacher and pupil STEM workshops and large scale STEM Fairs (both local and regional) with hands–on practical learning activities, career events and talks. They also manage national programmes such as the Nuffield Research Bursary Scheme and the British Science Association CREST Awards.
My children have all benefited enormously from a chance to work in science laboratories during the summer vacation, however there is the need to provide more placements, if you, your company or institution might be interested in helping please contact:-
STEM Team East 12 Ronald Rolph Court Wadloes Road Cambridge CB5 8PX http://www.stemteameast.org.uk
Molecular Interactions
I’ve just updated the molecular interactions page in the drug discovery resources, expanding the section on bonding to halogen.
Building a Screening Collection
I’ve just updated the page on building a screening collection and added links to recent useful publications.
Broad Coverage of Commercially Available Lead-like Screening Space with Fewer than 350,000 Compounds, Jonathan Baell DOI
Metal Impurities Cause False Positives in High-Throughput Screening Campaigns, Johannes C. Hermann et al DOI
Fragments in the Clinic Updated
Practical Fragments has updated the its list of fragment-derived compounds in the clinic. There is now one approved, the B-Raf(V600E) inhibitor Vemurafenib, eleven apparently in Phase 2 or 3 and fourteen reportedly in Phase 1.
Drug Discovery Resources Update
I’ve updated the Drug Discovery Resources Pages over the Christmas Break. In particular I’ve updated the Fragment Based Screening section and added a page on building a fragment collection. I’ve also updated the section on CYP interactions, expanding the Induction section.