Computational Chemistry Tools
Computational Chemistry can have a major impact on all stages of the drug discovery process, whether it be providing small desktop tools to enable scientists to access information more easily (See ChemSpider Safari Extension), calculation of physicochemical properties, virtual screening, structure-based design, QSAR analysis of both the desired target but also off-target activities.
There are a huge number of tools available, ranging in price from free to many $100,000s, here I’ll describe some of the tools I have used so whilst it is not comprehensive it might provide a useful stating point, the focus is on tools that a medicinal chemist might use.
See also Examples of Fingerprints and Descriptors.
ChemAxon
ChemAxon provide a number of desktop tools such as the drawing tool Marvin but also provide plugins to calculate a variety of physicochemical properties. In particular a tool to calculate pKa and the resulting LogD, probably the most important physicochemical property in medicinal chemistry. There are command line versions of these calculations that are invaluable for dealing with very large data-sets, these were used in a script in the analysis of fragment collections.
![]()
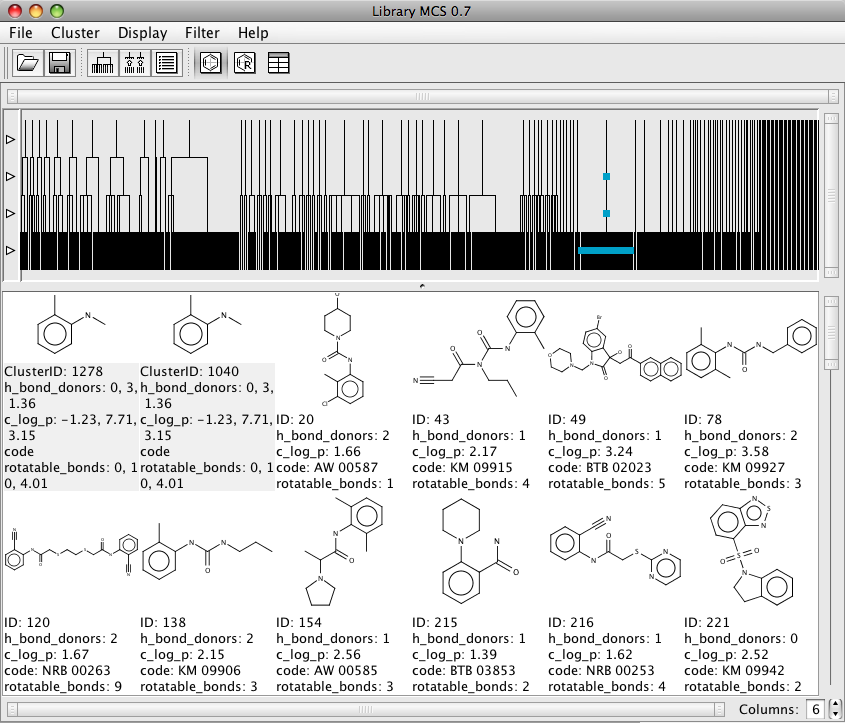
I’ve written a review of Marvin here. LibraryMCS is a tool from ChemAxon that uses hierachial clustering to sort molecules based on maximum common substructure, I find this an invaluable tool for looking at HTS data and is complimentary to other descriptor based clustering tools, an example can be found on the analysis of the Malaria screening.
ForgeV10
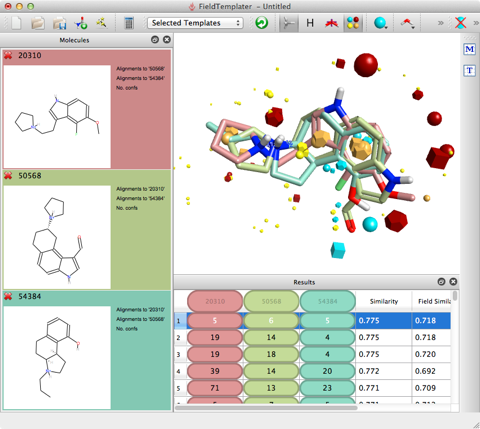
ForgeV10 allows the scientist to use Cresset’s proprietary electrostatic and physicochemical fields to align, score and compare diverse molecules. It allows the user to build field based pharmacophores to understand structure activity and then use the template to undertake a virtual screen to identify novel scaffolds.
Chemical Computing Group
MOE (Molecular Operating Environment) is my main Computational Chemistry tool and it provides nearly all the functionality required for drug discovery. MOE is a software system designed to support Cheminformatics, Molecular Modelling, Bioinformatics, Virtual Screening, Structure-based-design. I’ve written a couple of reviews of MOE here.
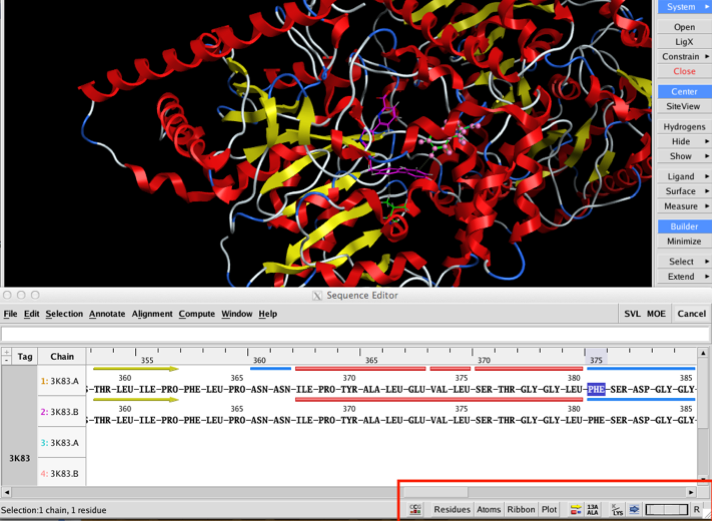
In addition the SVL scripting language also provides a powerful means to extend the capabilities of MOE and to build new tools. SVL Exchange, a repository of programs and code samples written in the Scientific Vector Language (SVL) by users and developers of the Molecular Operating Environment. The SVL Exchange is a valuable resource for users seeking specialized applications, utilities, or customizations for MOE. For users who program in SVL, the files in the SVL Exchange can also serve as examples of SVL programming. An example of the use of SVL can be found on the analysis of the Malaria screening where the two result sets were compared using a SVL script (dbnbmols_incommon) that identifies identical compounds in multiple databases, showing there were 49 identical compounds in the two results sets.
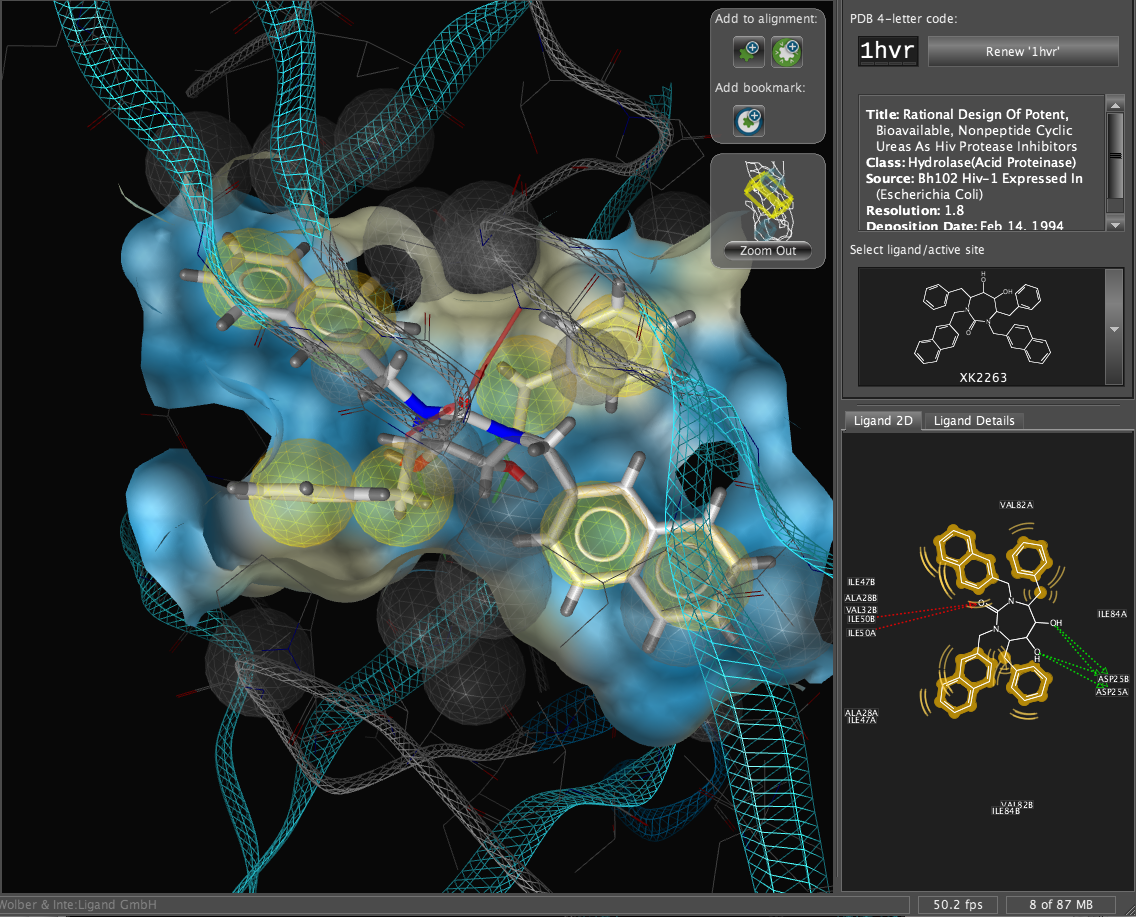
LigandScout
LigandScout from inte:ligand is a software tool that allows to rapidly and transparently derive 3D pharmacophores from structural data of macromolecule/ligand complexes in a fully automated and convenient way. There is a review here.
StarDrop
StarDrop is an application from Optibrium that was designed to aid decision making for scientists involved in drug discovery. StarDrop is a chemically aware data analysis package that allows you to quickly explore structure activity. It is easy to create multiple plots to compare data, and selection in one plot (in the image below the most active NK2 ligands were selected) automatically selects the corresponding molecules in all other plots and in the molecular spreadsheet. StarDrop comes with a variety of physicochemical property descriptor calculations and several ADME models are also available (Brain penetration, HERG and human intestinal absorption).
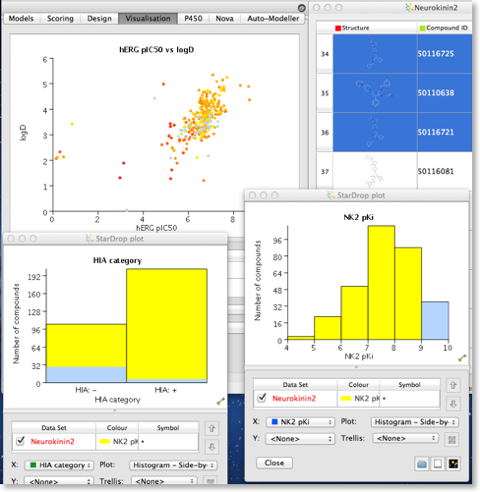
There are now links to external applications via plugins that link to Derek Nexus (Toxicity prediction) and Torch3D (Cresset’s field-based searching).
Vortex
Vortex is a chemically aware data analysis and spreadsheet tool from Dotmatics. You can import files or from a SQL database and do substructure or structural similarity searches. Calculate many physicochemical properties and perform data analysis and display.
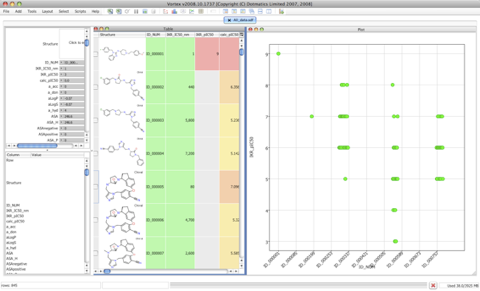
The ability to have multiple interactive plots of the data alongside grids of the highlighted structures is an enormous aid to understanding the data. All plots are linked such that selection in one plot is automatically selected in all other plots and spreadsheets.
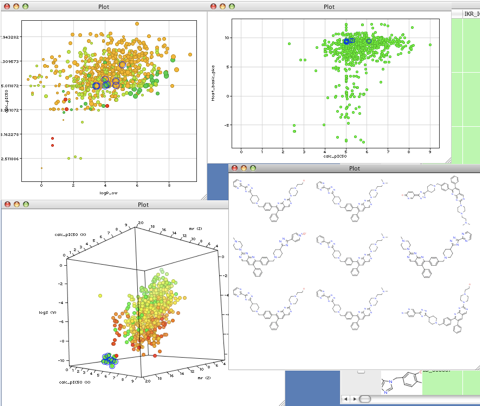
One of the very attractive features of Vortex is the availability of scripts that extend the capabilities of the applications. These can be used to extend the number of chemical descriptors available and also link to molecular modelling programs like MOE, statistical packages like R, or search and import information from databases and interact with web services.
FAME
FAME DOI is a collection of random forest models trained on a comprehensive and highly diverse data set of 20,000 small molecules annotated with their experimentally determined sites of metabolism taken from multiple species (rat, dog and human). In addition dedicated models are available to predict sites of metabolism of phase I and II processes.
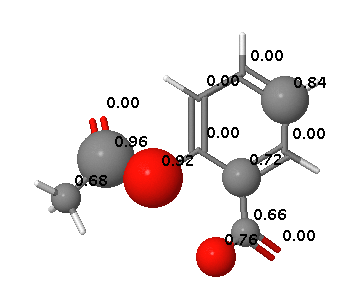
FAME offers a high performance prediction of sites of metabolism mediated by a wide variety of mechanisms.
The full review is available here
IMPACTS
IMPACTS (In-silico Metabolism Prediction by Activated Cytochromes and Transition States) is a hybrid site-of-metabolism (SoM) identification tool which combines docking to CYP enzymes, transition state (TS) modeling, and rule-based substrate reactivity prediction to predict the SoM of xenobiotics. The input is a 3D structure with the correct protonation state, Molecular Forecaster have their own drawing tools but since the file format is .mol2 these could be generated using a variety of alternative tools. The output includes the structure bound to the CYP and the putative metabolites.
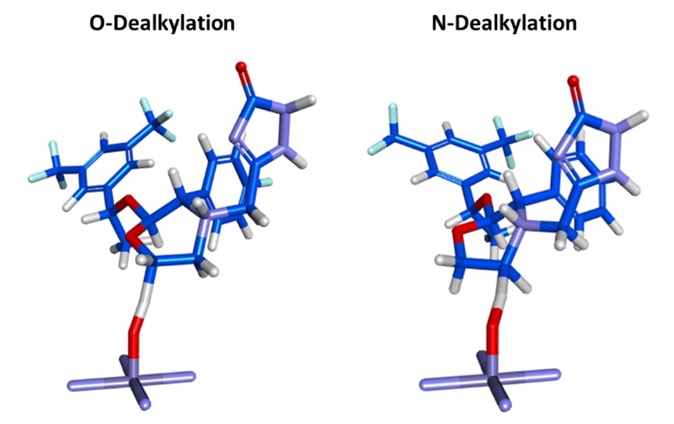
Campagna-Slater V., Pottel J., Therrien E., Cantin L.-D., Moitessier N., J Chem Inf Model, 2012, 52, 2471-2483 DOI
Open Source tools
There are now a variety of very useful open source tools that can be used for drug discovery. The Royal Society of Chemistry Chemical Information and Computer Applications Group have run a series of 20 workshops highlighting different tools, these are all now freely available on YouTube https://www.youtube.com/c/RSCCICAG.
Listing of applications by function
GUI
There are some suites of tools that provide almost all of the functionality of the applications described below in a single unified interface.
MOE, Sybyl, Discovery Studio, ICM Pro
Molecule Viewers
These are used to view the 3-dimensional structure of a molecule, many are also capable of displaying biological macro,molecules such as proteins.
PyMol, VIDA, Chem3D, Chimera, Jmol, JSmol, AstexViewer, CN3D, CylView, DINO, ICM Browser. Molgro Viewer, Qutemol, VMD, Yasara, ICM Browser
Chemical Sketchers
These are small molecule drawing packages that can be used to create the input for other programs, rendering structures on web pages, and some also provide publication quality output. Traditionally these were heavy duty desktop packages however with the increase in the use of web-based tools there are now a variety of lightweight javascript based sketchers. Many of these new lightweight tools have the advantage in that they can be used on mobile devices like an iPad or smart phone.
ChemDraw, Marvin, JME, ChemDoodle, Elemental, JSDraw, PubChem Sketcher, Ketcher
SMARTSviewer generates a visualisation of a molecular pattern that is given in form of a SMARTS very useful for checking that your search query is what you really want.
Chemical Property Calculations
Marvin, Elemental, Stardrop, Vortex, PaDEL (see also Tookits below)
3D structure generation, conformers
Many drawing packages will generate 1D (SMILES) or 2D (sdf, mol) representations of molecules. However some of the virtual screening tools require 3D structures and often a selection of reasonable conformations.
OMEGA, CORINA, ROTATE, CONFLEX
Docking
Some docking applications will generate a 3D structure and multiple conformations of the proposed ligand, many also include a variety of scoring functions to rank the proposed poses.
Autodock, Dock, FlexX, Autodock Vina, Fred, Haddock, Bude, Zdock, GOLD, Glide, FITTED, FlipDock, ADAM, MS-Dock, UCSF Dock, ParDock, FlipDock, PLANTS
Similarity, shape or pharmacophore screening
Whilst docking is an important tool for the generation of novel ligands it should be remembered that searching using simple 2D ligand, pharmacophore or shape descriptors can offer excellent results in a fraction of the time that a docking run requires.
ROCS, LigandScout, Aligin-it, EON, Forge, Blaze, LibMCS, Open3DQSAR, Shape-it, GRID, PHASE, Catalyst
Data visualisation
There are a lot of data analysis and visualisation tools so here I’ve only highlighted those that come with chemical intelligence built in.
Vortex, StarDrop, Spotfire, Molgro Data Modeller, Sarchitect, Activity Miner, Instant JChem, Accord for Excel, DataWarrior
Workflow or automation
Many of the tools can be accessed by Python (particularly Jupyter Notebooks), Shell, or applescript however there are now specialist tools that allow the construction of workflows to simplify repetitive tasks.
KNIME, Pipeline pilot, Taverna
Toolkits
These allow you to build your own applications with the chemical intelligence provided by the toolkit, they can be accessed using C, C++, Java, Python, PERL etc. They will also have a command line interface. They can also be used to add chemical property calculations etc. to other applications.
OpenBabel, RDKit, Cactus, CDK, Digital Chemistry, OECHEM, Daylight, JChem, MayaChem tools, PerlMol
Web Resources
Dock Blaster, Haddock, DockingServer, GRAMM-X, SwissDock, iDock, TarFisDock
Project Management
As a project generates more and more data it becomes increasingly difficult to manage especially as results are often stored in excel tables or delimited text files. To efficiently store, manage and mine this information it is essential to have a informatics solution that allows all scientists access. This is especially true if science is being carried out on multiple sites. Whilst there are many enterprise level project management systems very few have any scientific intelligence, those mentioned below are founded in science. In addition the older enterprise systems seem to be rather inflexible systems, requiring significant desktop support, the newer systems simply require a web browser interface.
Dotmatics, Accelrys, ThremoScientific,PerkinElmer
Worth Reading
Open Source Molecular Modeling DOI a review that categorizes, enumerate, and describe available open source software packages for molecular modeling and computational chemistry.
There is also an online database https://opensourcemolecularmodeling.github.io that covers most aspects of computational drug discovery
Methods
Development Activity
Usage Activity
Cheminformatics
Toolkits
Standalone Programs
Graphical Development Environments
Visualization
2D Desktop Applications (Table [2ddesktopviz])
3D Desktop Applications
Web-Based Visualization
QSAR/ADMET Modeling
Descriptor Calculators
Model Building
Model Application
Visualization
Quantum Chemistry
Ab initio Calcuation
Helper Applications
Visualization
Ligand Dynamics and Free Energy Calculations
Simulation Software
Simulation Setup and Analysis
Virtual Screening and Ligand Design
Ligand-Based
Docking and Scoring
Pocket Detection
Ligand Design
Machine learning is a critical tool for drug discovery, it is used for predictive modelling in many areas. The choice of tool is often a personal preference but this paper is a good start for comparisons.
Do we Need Hundreds of Classifiers to Solve Real World Classification Problems
We evaluate 179 classifiers arising from 17 families (discriminant analysis, Bayesian, neural networks, support vector machines, decision trees, rule-based classifiers, boosting, bagging, stacking, random forests and other ensembles, generalized linear models, nearest-neighbors, partial least squares and principal component regression, logistic and multinomial regression, multiple adaptive regression splines and other methods), implemented in Weka, R (with and without the caret package), C and Matlab, including all the relevant classifiers available today. We use 121 data sets, which represent the whole UCI data base (excluding the large- scale problems) and other own real problems, in order to achieve significant conclusions about the classifier behavior, not dependent on the data set collection. The classifiers most likely to be the bests are the random forest (RF) versions, the best of which (implemented in R and accessed via caret) achieves 94.1% of the maximum accuracy overcoming 90% in the 84.3% of the data sets. However, the difference is not statistically significant with the second best, the SVM with Gaussian kernel implemented in C using LibSVM, which achieves 92.3% of the maximum accuracy. A few models are clearly better than the remaining ones: random forest, SVM with Gaussian and polynomial kernels, extreme learning machine with Gaussian kernel, C5.0 and avNNet (a committee of multi-layer perceptrons implemented in R with the caret package). The random forest is clearly the best family of classifiers (3 out of 5 bests classifiers are RF), followed by SVM (4 classifiers in the top-10), neural networks and boosting ensembles (5 and 3 members in the top-20, respectively).
Clustering is an invaluable cheminformatics technique for subdividing a typically large compound collection into small groups of similar compounds. One of the advantages is that once clustered you can store the cluster identifiers and then refer to them later this is particularly valuable when dealing with very large datasets. This often used in the analysis of high-throughput screening results, or the analysis of virtual screening or docking studies. This review looked at a number of options for clustering molecules from toolkits like RDKit to commercial applications such as Vortex and tried them with a variety of different sized data sets, one containing 789 molecules, one 150,000 molecules and a large set containing 4,400,000 molecules.
See also Examples of Fingerprints and Descriptors
Last Updated 8 December 2022