Fragment-Based Screening Published Hits
Whilst there are a variety of techniques to measure the properties or diversity of fragment libraries I thought it might be interesting to look at the profiles of compounds that actually appear as hits in fragment-based screening campaigns. I’ve been compiling a database, for several years, of fragments that have been reported as hits in the literature, this database now has over 1500 entries culled from over 310 publications directed at nearly 220 different molecular targets using 26 different detection technologies and might be expected to give some insight into the type of compounds that appear as hits. With the caveat that the dataset only includes information that has been published.
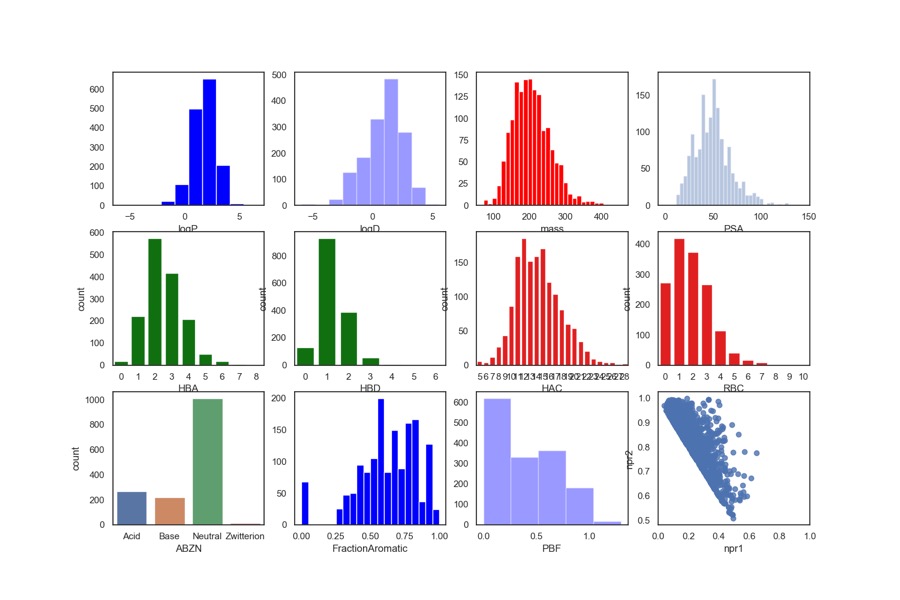
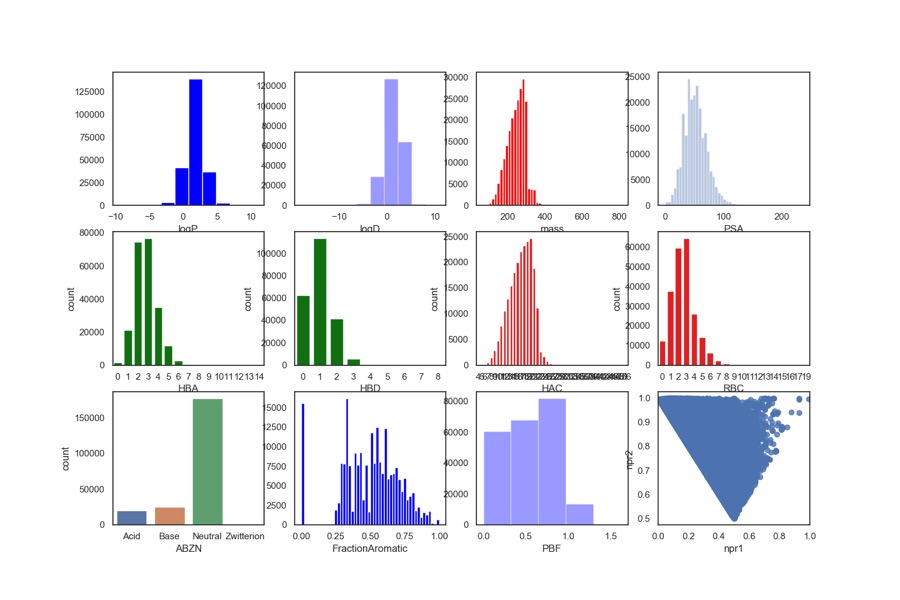
Originally the physicochemical properties were calculated using a combination of AppleScript, ChemAxon and Aabel but that has now been replaced by a Jupyter Notebook in which we can monitor and automate the entire workflow. The Jupyter notebook can be downloaded from here together with a detailed description. It uses the command line tool Evaluate from ChemAxon to calculate (HBD, HBA, PSA, HAC, LogP, LogD, MWt, RBC), Evaluate was used to determine pKa in order to identify acidic or basic groups and categorized the fragments accordingly (Acidic, Basic, Neutral or zwitterion). The fraction of aromatic atoms (number of aromatic atoms/number of heavy atoms) was also included using the ChemAxon aromaticity model. I’ve also included npri (Normalized ratio of principle moments of inertia) as described by Sauer WH, Schwarz MK (2003) Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J Chem Inf Comput Sci 43:987–10030. DOI this was calculated using MOE. I’ve recently added “Plane of best fit” (A Novel Method to Characterize the Three-Dimensionality of Molecules, Nicholas C. Firth, Nathan Brown, and Julian Blagg, Journal of Chemical Information and Modeling 2012 52 (10), 2516-252 DOI. ) The plots were then generated using a Python data visualization library based on matplotlib Seaborn.
The results are shown in the collection of histograms below, in general the fragments are within the rule of three guidelines. One notable observation is that around 1/3 of the hits contain an ionisable group (pKa predicted using ChemAxon tools) and this has an impact when comparing cLogP and cLogD. Whilst the mean LogP is around 1.7, the mean LogD is around 1.5. The molecular weight is almost invariably below 300 with the majority below 250, with the heavy atom count (HAC) 16 or lower. The lack of rotatable bonds (RBC) and the relative high Fraction Aromatic scores are consistent with the observation that many of the hits are substituted aromatic or heteroaromatic rings, in fact only 297 of the 1503 fragment structures contain no aromatic atoms. Looking at the npri plot the compounds are clustered along the axis between rod-like and disc-like again supporting the view that the hits have limited 3D shape. The plane of best fit data also underlines the paucity of 3D structures.
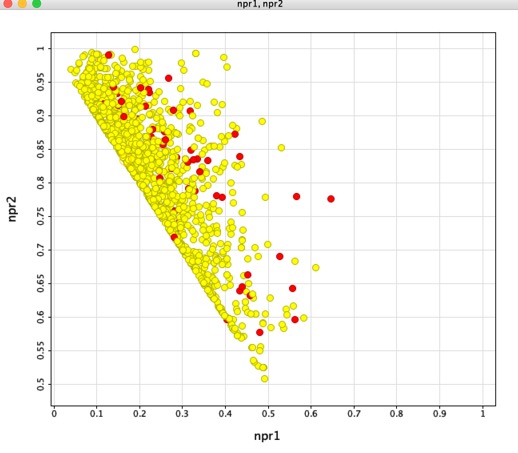
Looking at the npri plot the compounds are clustered along the axis between rod-like and disc-like again supporting the view that the hits have limited 3D shape. The plane of best fit data also underlines the paucity of 3D structures. In the plot of npr1 versus npr2 shown below, the fragments containing no aromatic atoms are highlighted in red, the results suggest, unsurprisingly, that they do perhaps explore more 3D space.
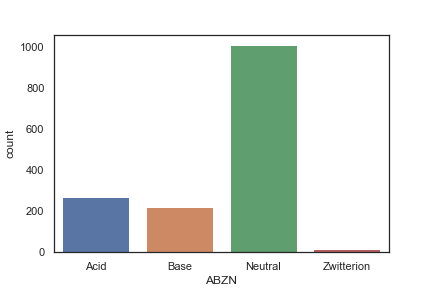
Approximately one third of the fragments are predicted to be ionised at physiological pH, this could be due to the need for high solubility in the fragment screen, and/or it could be due to the ionised group providing a key binding interaction.
The calculated physicochemical properties profile needs to be taken with a pinch of salt since you can only test what is available, however a comparison with the corresponding physicochemical profile of all available fragments in commercial fragment collections (approx 250,000 fragments) does highlight some interesting differences. The mean LogP/D for the available fragments is actually lower than the published fragments.
Perhaps the most notable difference is that the proportion of ionisable groups in the published fragments is much higher than found in the available fragments, this could reflect the need for a strong interaction to aid identification of fragment binding or it could be that improved solubility enables screening at higher concentrations. In addition the published fragments tend be of lower molecular weight and have a greater proportion of aromatic atoms. Whilst there are a significant number of fragments with greater 3D character it would appear that this is not reflected in the published fragments.
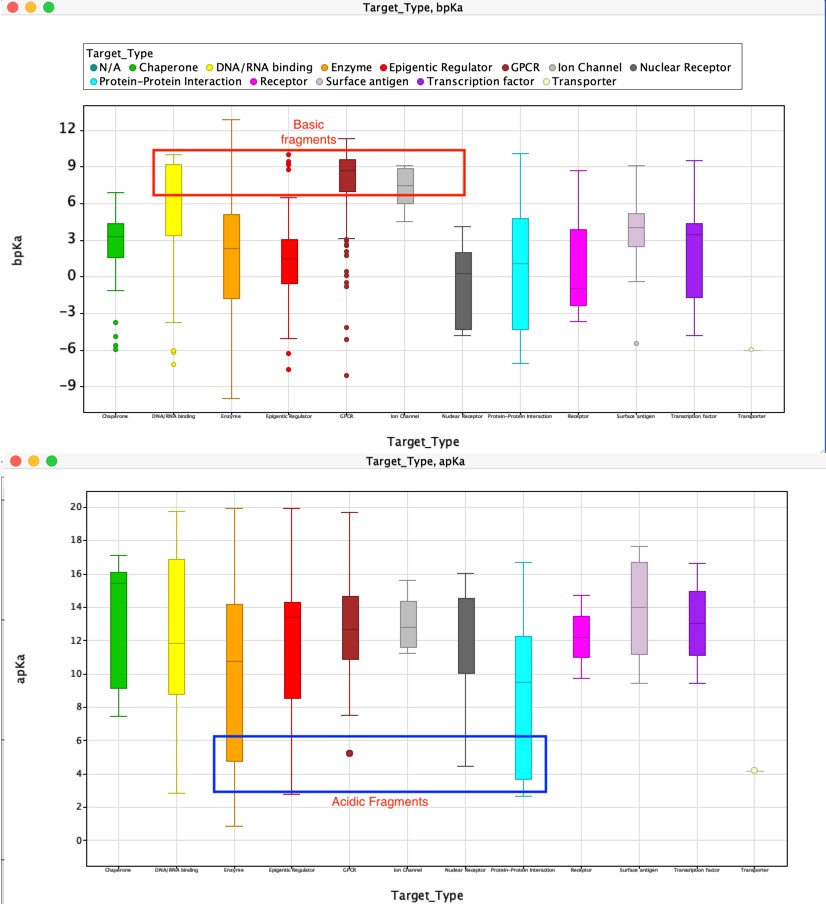
In general the calculated physicochemical properties are similar across all the Target Classes, the exception being pKa. As the box plot below illustrates DNA binding, GPCR and ion channels appear to favour basic ligands, whilst protein-protein interactions and some enzymes favour acidic fragments.
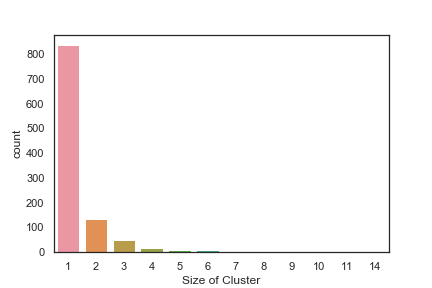
I also clustered the compounds (Using Morgan fingerprints) within the Jupyter notebook and the results are shown in the histogram below, whilst the majority are singletons there are a number of 2, or 4- membered clusters, although it has t be said that fragments will generate rather sparse fingerprints. However, closer inspection shows these clusters are in fact identical compounds that have been identified as hits in different fragment screening campaigns against different targets.
Functional Group Analysis
I used Checkmol to analyse the fragments for the presence of various functional groups and structural elements. Selected results are shown below.
- 1437/1505 contain an aromatic ring, 1202 of which are heteroaromatic
- 332 contain an arylhalide, 162 contain a phenol
- 267 contain an acidic group, 218 a basic group
- 30 contain a nitro group
- 178 contain a hydroxy, 126 an ether
- 598 contain an amine, 275 “anilines” (mainly on heteroaromatic systems)
- 224amides, 55 esters, 31 ureas
Bonds to halogen are similar to hydrogen bonds and there are many examples in the PDB of carbonyls interacting with halogens with bonds to I, and Br predominating. It is perhaps worth noting that halogens have been introduced into ligands to aid NMR or X-ray analysis, but they may also influence binding. Based on the observed bond angles the interaction is between the halogen and pi-cloud of the carbonyl rather than the lone pair with a clear clustering of X--O=C-N dihedral angles of 90° associated with interactions that involve primarily the pi-system of the carbonyl. Although these interactions may be thought of a weaker than hydrogen bonds you should bear in mind the impact on desolvation of the hydrogen bonding partners may have on the overall energy change. Desolvation of the halogen may be less of an issue and so the overall benefit may be higher.
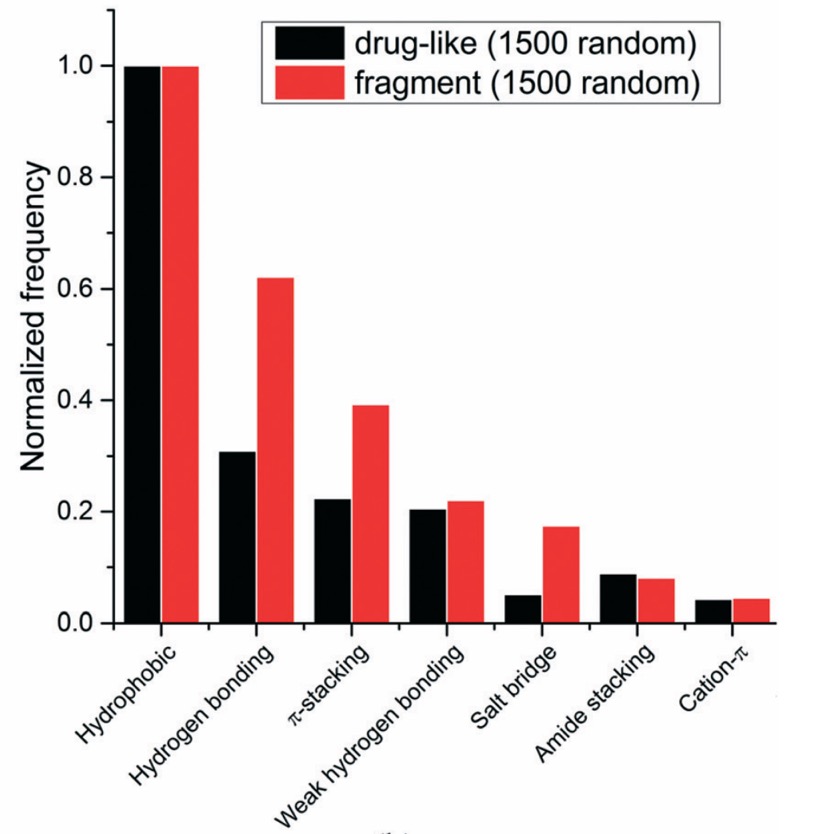
A detailed analysis of the molecular interactions present found between ligands and macromolecules has been undertaken, "A systematic analysis of atomic protein–ligand interactions in the PDB" DOI looking at over 11,000 complexes the authors were able to categorise the 7 most common types of interaction and compare drug-like sized molecules with fragments. Interestingly Hydrogen bonds, π-stacking interactions and salt bridges are more common among the fragment ligands, which is consistent with the high proportion of aromatic rings and ionisable groups found in the published fragment hits.
Most Common Scaffolds
To identify the most common scaffolds I used sca.svl with MOE. The script finds all scaffold in a database, writes them to a separate database. Interestingly the top 10 scaffolds are all aromatic systems.
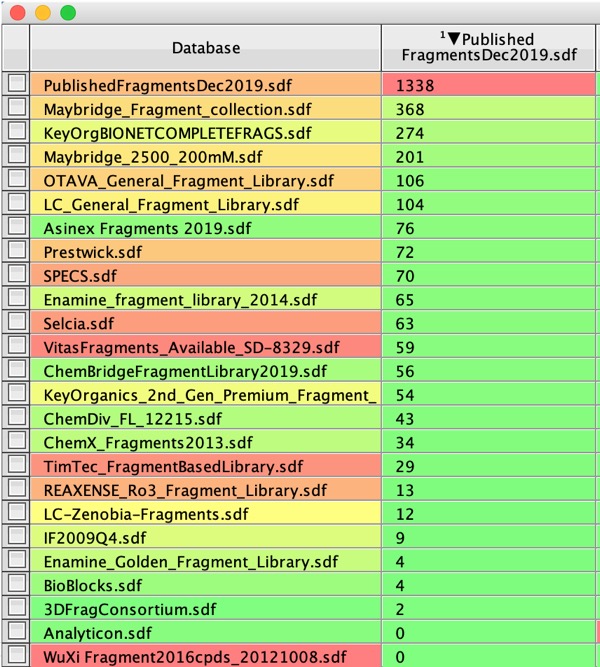
Suppliers
I’ve also looked to see which of the published fragments are in the commercial fragment collections, the results are shown in the table below. The Vortex script compares InChIkeys between the databases (there are 1338 unique InChiKeys in the Published Fragments dataset because there are multiple duplicate fragments that have been identified in different screening campaigns).
It is clear that some collections contain many more of the published hits than others, I suspect that this reflects those vendors who were the first to determine the solubility od the fragment collections.
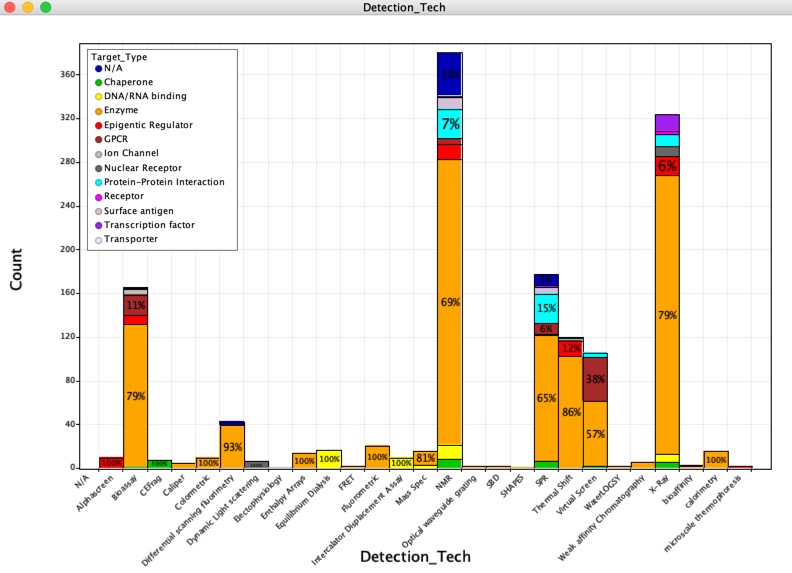
Detection Technology
A total of 26 different technologies have been employed and whilst NMR and X-ray have been the historical preferred technologies, thermal shift has become more common recently. It seems that on many occasions thermal shift or SPR are used for a rapid screen and then X-ray or NMR are used to confirm hits. Colour coding by target type highlights GPCR targets (brown) are most often screened either by virtual screening or bioassay, as might be expected for membrane bound proteins.
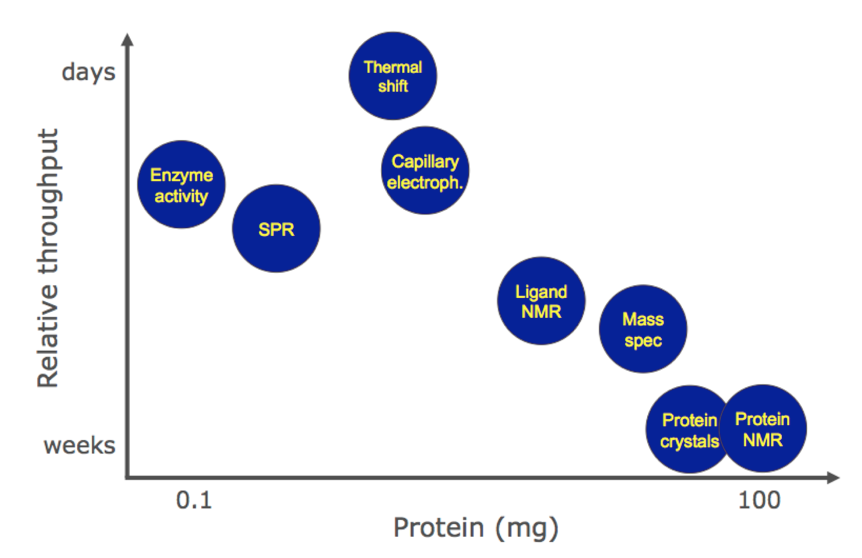
The increasing popularity of thermal shift is perhaps because this fragment-screening technologies offer the best compromise between speed of throughput and protein requirements. However to drive structure-based design projects structural information provided by NMR or X-Ray is critical.
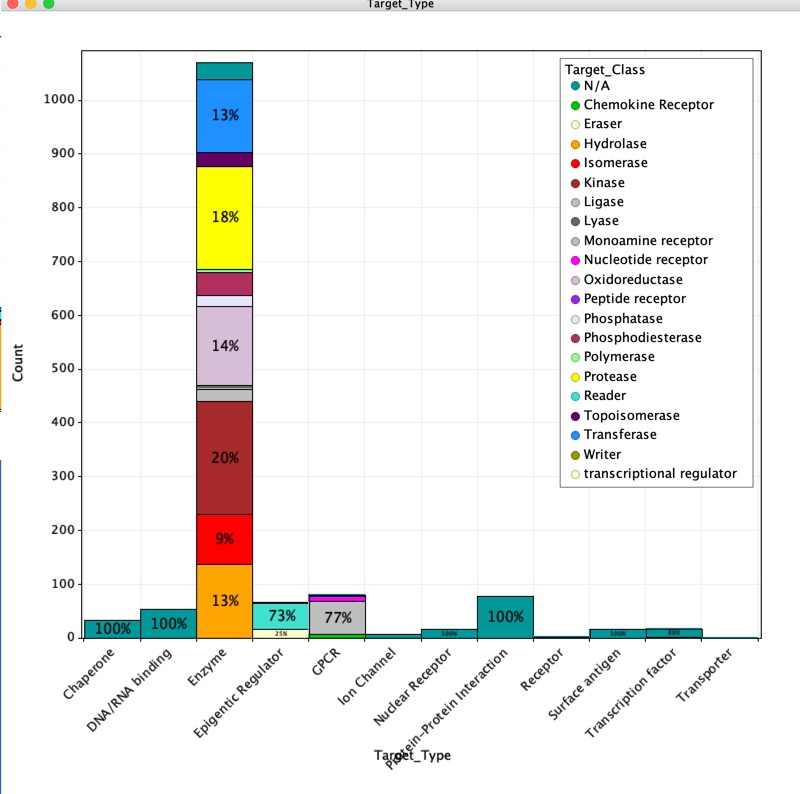
Target Types
For each reference the target description is captured and used to identify the Uniprot ID, this was then used to search ChEMBL to add the target type/class description, using ChEMBL ontology. Where available the affinity and how it was determined was also included. If a crystal structure was disclosed either as part of the fragment screen, or to confirm a fragment hit identified using another screening technology, the PDB code was added to the database.
A plot of the different target types is shown below, as might be expected Enzymes predominate, with Kinases and Proteases making a major contribution. However, a wide variety of target types have now been addressed using fragment based screening approaches.
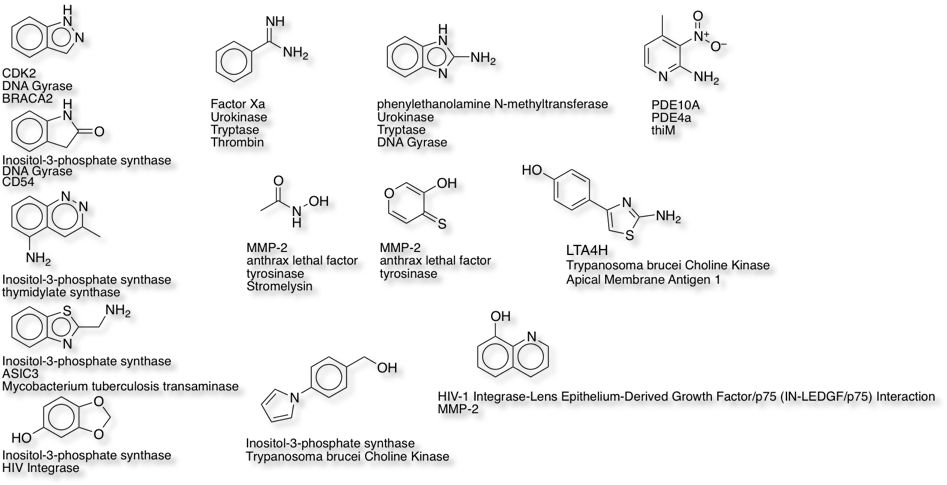
Interestingly, as highlighted in the clustering above, there are a number of fragments that have been identified for multiple targets, and it is notable that many are active against seemingly unrelated proteins. Whilst there might be concern about promiscuous ligands, inspection of the available X-ray structures suggests these fragments form specific binding interactions.
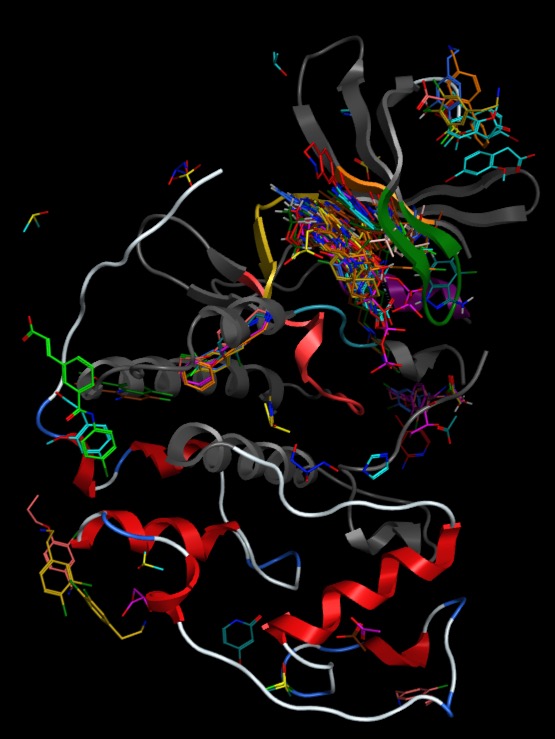
Of the 210 fragment hits identified for kinase targets the crystal structures for 69 are in the PDB. These were downloaded using a Jupyter Notebook and then imported into MOE. The structures were superimposed using the hinge region as the target and then the all but one of the proteins hidden. This ribbon structure was then colour coded using the kinase annotation. The vast majority appear to bind to the hinge region.
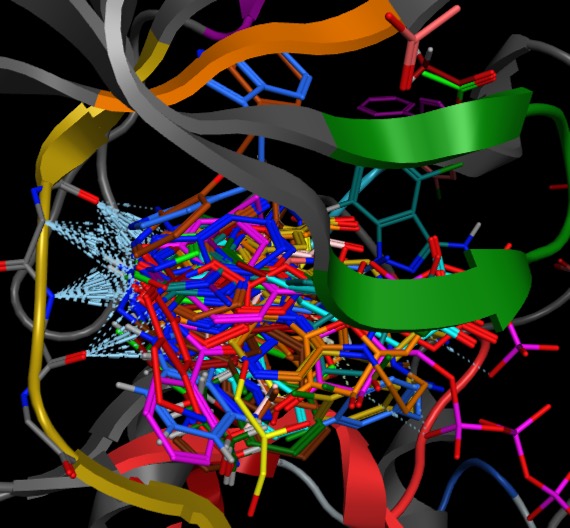
With the key hydrogen bonding interactions between fragments and the hinge region (coloured yellow) highlighted below.
Similarly, 1-methylquinolin 2-one binds to both the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) and the human PCAF bromodomain in an identical manner as shown below (PDB codes 5FE1 and 4QST).
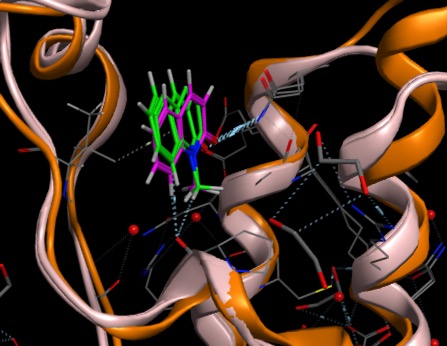
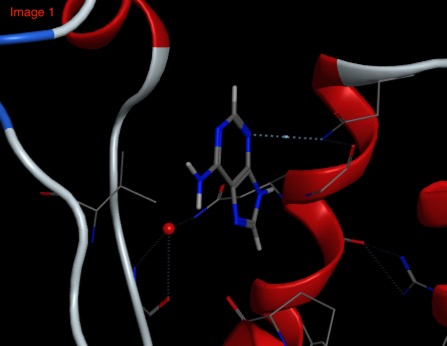
In contrast whilst Adenine binds in a similar to the BAZ2B Bromodomain (Image 1 below) as the ligands above with the aromatic sitting over a Valine side-chain and N3 hydrogen bonds to an Asparagine, when adenine binds to PNMT (phenylethanolamine N-methyltransferase) Image 2 below) the aromatic system sits over a phenylalanine and N3 hydrogen bonds to solvent.
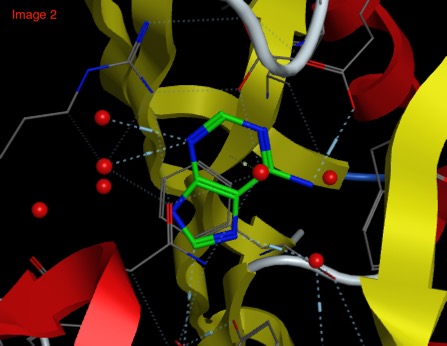
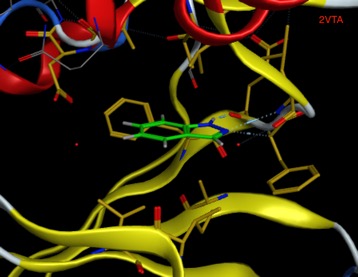
In another example the fragment Indazole binds to CDK (2VTA below) in a face-to-face manner with a beta-sheet, whilst in RadA (4B2I) it bind in an edge-to-face manner with a beta-sheet.
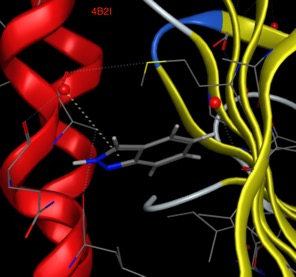
It seems that fragments can adopt a variety of binding poses.
How well do they bind?
As you might expect fragments have modest/weak affinity, this is not measured for all examples but the vast majority are in the range 1-1000 uM. A number of different technologies have been used to measure affinity but bioassay data is the most commonly reported.
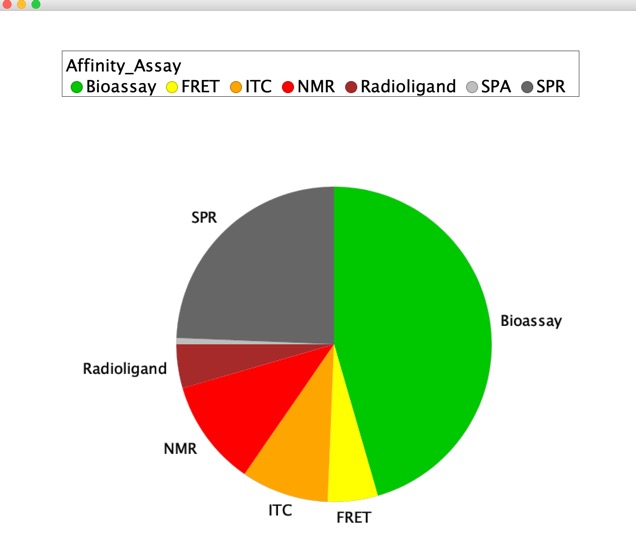
Detection Technology
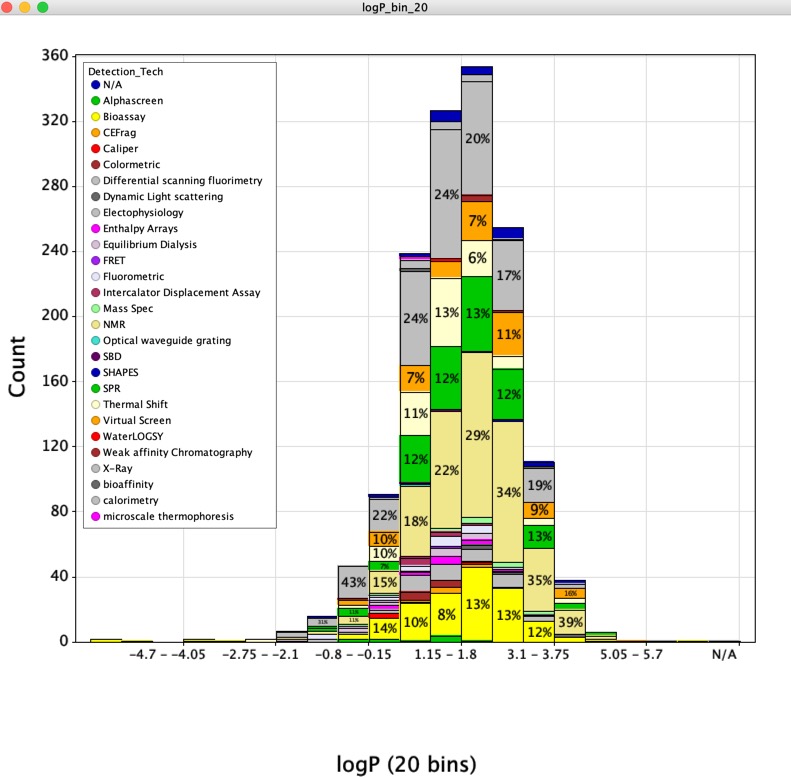
Evidence from literature that different technologies can identify the same hits for a single target. No evidence that detection technology influences the physiochemical properties of the hits identified (LogP shown below). Some technologies (e.g. SPR) are thought to have a higher false positive rate.
How Are Fragments Optimised?
A recent paper J. Med. Chem. 2013, 56, 2478−2486 DOI looked at the different ways that the initial fragments were subsequently optimised, looking at a variety of physicochemical properties and ligand efficiency indices, in particular they emphasise the need to use size independent ligand efficiency measures. Lipophilicity metrics, logP and LELP were found to be particularly useful as all clinical candidates arose from fragment-based programs were located in, or near to the logP 0−5 and LELP 0−10 space.
LELP = (log P / LE)
Several conclusions were then drawn.
Hit detection methods were found to affect hit quality and biochemical hits were found to be especially advantageous. Biochemical hits not only exhibit better affinity and lipophilicity indices over NMR and X-ray hits, but compounds optimized from them preserve their advantageous properties over those optimized primarily from NMR and partly from X-ray hits.
It appears that the use of atomic level structural information of the target (most often from X-ray but also from NMR) advantageously influences compound properties.
The dataset includes work from universities (18%), Large Pharma (45%) and small/medium pharma (37%), with the small medium pharma producing results that were superior to Large Pharma and universities, probably because protein structural information plays a critical role in the small/medium pharma companies.
Whilst getting fragment hits has now become a relatively predictable process A general consensus from scientists working in the area is that the key to successful prosecution of fragment-based drug design is the availability of structural information, so whilst there are a steadily increasing number of technologies available for the initial screening of fragments generating the protein crystal structure is still a critical and often rate-limiting step in the process. Diamond provide a tips and tricks page based on their (and SGC) experience Tips and tricks. It might also be worth reading "High-throughput production of human proteins for crystallization: The SGC experience" DOI and "Lessons from high-throughput protein crystallization screening: 10 years of practical experience" DOI.
This paper is worth reading "Binding-Site Compatible Fragment Growing Applied to the Design of β2-Adrenergic Receptor Ligands" DOI, it describes a workflow aimed at identifying fragment derivatives that can be prepared using a suite of robust chemical reactions.
In order to address the synthetic tractability issue, our in silico workflow aims at derivatized products based on robust organic reactions. The study started from the predicted binding modes of five fragments. We suggested a total of eight diverse extensions that were easily synthesized, and further assays showed that four products had an improved affinity (up to 40-fold) compared to their respective initial fragment. The described workflow, which we call “growing via merging” and for which the key tools are available online, can improve early fragment-based drug discovery projects, making it a useful creative tool for medicinal chemists during structure–activity relationship (SAR) studies
Updated 5 January 2020