ChEMBL 35 is out
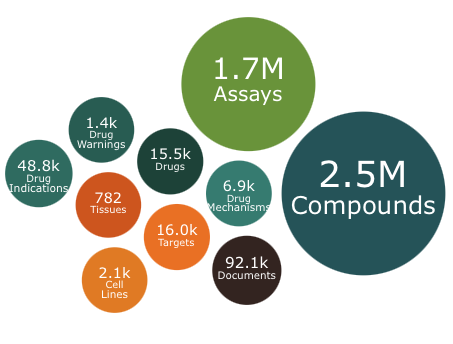
The year ends with an update to ChEMBL. This release contains 2.5 million compounds and 1.7 million assays including over 15K drugs or molecules in development.
You can download the dataset in various formats https://chembl.gitbook.io/chembl-interface-documentation/downloads.
Full details of the update are on the ChEMBL blog. https://chembl.blogspot.com/2024/12/heres-nice-christmas-gift-chembl-35-is.html.
SureChEMBL Updated
SureChEMBL is a database of automatically abstracted patents, it uses three different methods to get structures, name to structure, image to structure and for some patents mol files if available. If you use it regularly you will be aware that it has become somewhat unreliable and the performance is not ideal.
This has just changed with an updated version of SureChEMBL.
Almost 10 years ago, EMBL-EBI acquired the SureChem system of chemically annotated patents and made this freely accessible in the public domain as SureChEMBL. Since then, our team has continued to maintain and deliver SureChEMBL. However, this has become increasingly challenging due to the complexities of the underlying codebase. We were awarded a Wellcome Trust grant in 2021 to completely overhaul SureChEMBL, with a new UI, backend infrastructure, and new features. We are now able to make available the first outputs from this project, which addresses the first two of these deliverables, with more to come in the future!
The new interface is here https://www.surechembl.org
If you have any issues you can submit them on GitHub https://github.com/chembl/surechembl-issues/issues.
One particularly useful new feature is the new public api. https://www.surechembl.org/api/swagger-ui.html. I'll certainly be exploring this in the future.
ChEMBL 33 released
The latest update to ChEMBL has been released.
This fresh release comes with a few new data soures and also some new features: we added bioactivity data for understudied SLC targets from the RESOLUTE project and included a flag for Natural Products and for Chemical Probes. An annotation for the ACTIONTYPE of a measurement was included for approx. 270 K bioactivities. We also time-stamped every document in ChEMBL with their CREATIONDATE!
This version of the database, prepared on 31/05/2023 contains:
2,399,743 compounds (of which 2,372,674 have mol files)
3,051,613 compound records (non-unique compounds)
20,334,684 activities
1,610,596 assays
15,398 targets
88,630 documents
Full details are here http://chembl.blogspot.com/2023/06/release-of-chembl-33.html.
ChEMBL is a manually curated database of bioactive molecules with drug-like properties. It brings together chemical, bioactivity and genomic data to aid the translation of genomic information into effective new drugs.
Fake Publications in Biomedical Science
There have a number of headlines recently highlighting large language models (LLM https://en.wikipedia.org/wiki/Largelanguagemodel, most notably GTP-4 from OpenAI. These models are trained on vast amounts of data from a variety of sources and the quality of these data sources is not always as good as hoped.
It might be assumed the scientific literature would be of a higher standard but a recent preprint raises major concerns.
https://www.medrxiv.org/content/10.1101/2023.05.06.23289563v1
Fake Publications in Biomedical Science: Red-flagging Method Indicates Mass Production
Red-flagged fake publications (RFPs) account for around 28% of the published papers in biomedicine.
ChEMBL 32 is released!
The fantastic resource ChEMBL has been updated. ChEMBL 32 contains
- 2,354,965 compounds (of which 2,327,928 have mol files)
- 2,995,433 compound records (non-unique compounds)
- 20,038,828 activities
- 1,536,903 assays
- 15,139 targets
- 86,364 documents
More details are here http://chembl.blogspot.com/2023/03/chembl-32-is-released.html.
Data can be downloaded from the ChEMBL FTP site.
ChEMBL 31 is released
The latest release of the absolutely invaluable ChEMBL database is available.
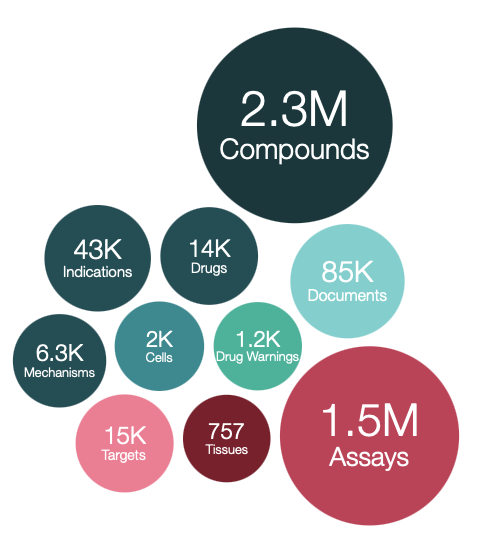
This version of the database, prepared on 12/07/2022 contains:
2,967,627 compound records
2,331,700 compounds (of which 2,304,875 have mol files)
19,780,369 activities
1,498,681 assays
15,072 targets
85,431 documents
Available from the downloads page https://chembl.gitbook.io/chembl-interface-documentation/downloads
ChEMBL 28 has been released.
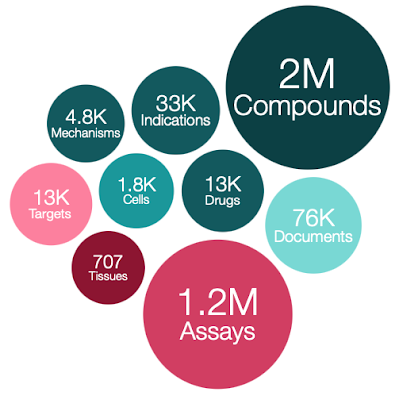
I see that a new version of ChEMBL has been released. Chembl 28
- 2,680,904 compound records
- 2,086,898 compounds (of which 2,066,376 have mol files)
- 17,276,334 activities
- 1,358,549 assays
- 14,347 targets
- 80,480 documents
ChEMBL 26 Released
The latest release of the essential molecule bioactivity dataset has just been announced.
ChEMBL 26 contains
- 2,425,876 compound records
- 1,950,765 compounds (of which 1,940,733 have mol files)
- 15,996,368 activities
- 1,221,311 assays
- 13,377 targets
- 76,076 documents
A couple of notes
We are now using RDKit for almost all of our compound-related processing. For the first time in ChEMBL26, this will include compound standardization, salt-stripping, generation of canonical smiles, structural alerts, image depiction, substructure searches and similarity searches (via FPSim2: https://github.com/chembl/FPSim2). Therefore, all molecules have been reprocessed and you may notice some differences in molfiles, smiles and structure search results compared with previous releases. The ChEMBL structure curation pipeline has been released as an open source package: https://github.com/chembl/ChEMBLStructure_Pipeline, and incorporated into our Beaker web services (see below). More information can be found here: http://chembl.blogspot.com/2020/02/chembl-compound-curation-pipeline.html.
We are also now using ChemAxon tools to calculate most acidic and basic pKa, logP and logD (pH 7.4) predictions, rather than ACDLabs software. These properties have therefore been recalculated and renamed in the database.
The release notes contain more details and the database can be downloaded from the ChEMBL FTP site.
Neglected and Tropical Diseases
I've tried to support research in the Neglected Tropical Disease area in several ways, I organised a session at the 19th Cambridge MedChem Meeting in 2017 and arranged for the session to be recorded and is now available online and has been watched nearly 300 times.
This is a recording of the Neglected and Tropical Diseases Session at the 19th Cambridge MedChem Meeting, 11-13 September 2017. The speakers are Kelly Chibale (Univ of Capetown), Christoph Boss (Actelion), Rob Young (GlaxoSmithKline), Jonathan Large (LifeArc) and Charles Mowbray (DNDI).
So I was delighted to hear that IUPHAR/BPS Guide to pharmacology database have been funded by MMV to add details of antimalarials to their database.
This link http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=970 gives details of antimalarial targets, including gene name, synonyms and Uniprot ID.
This link http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=999 gives details of antimalarial ligands, including mode of action and properties. For example artemisinin.
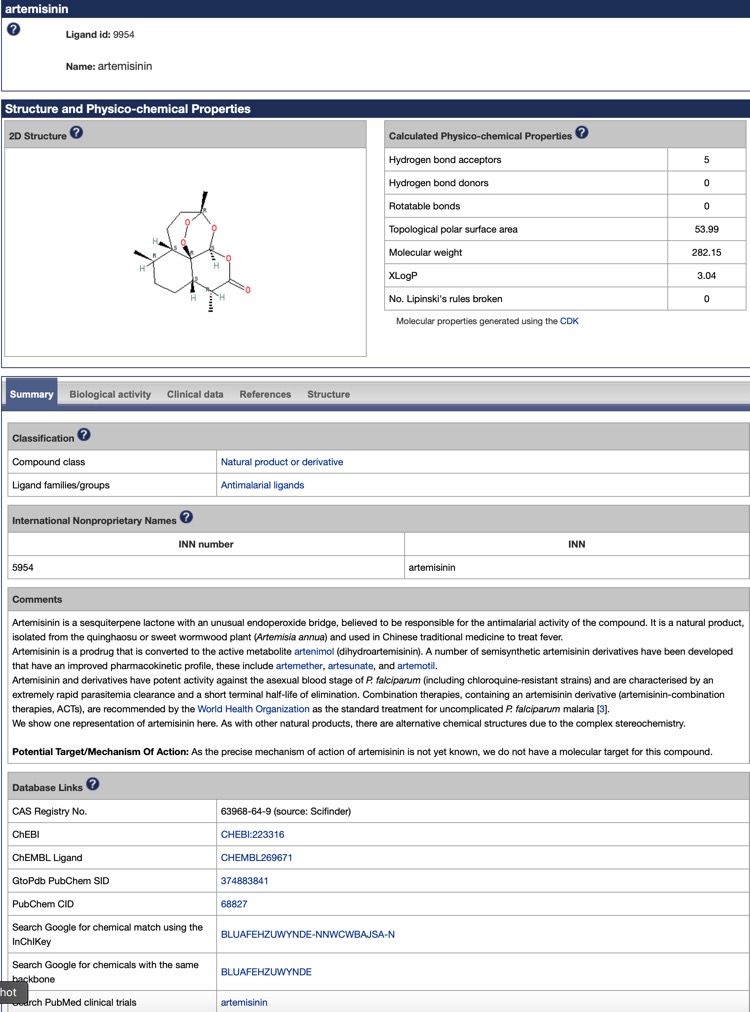
I'd urge you have a look and I'm sure they would be happy to hear any suggestions.
ReFRAME library as a comprehensive drug repurposing library
This looks a very interesting resource described in a recent publication. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis DOI.
The ReFRAME collection of 12,000 compounds is a best-in-class drug repurposing library containing nearly all small molecules that have reached clinical development or undergone significant preclinical profiling. The purpose of such a screening collection is to enable rapid testing of compounds with demonstrated safety profiles in new indications, such as neglected or rare diseases, where there is less commercial motivation for expensive research and development.
To date, 12,000 compounds (80% of compounds identified from data mining) have been purchased or synthesized and subsequently plated for screening. In addition, an open-access data portal (https://reframedb.org) has been developed to share ReFRAME screen hits to encourage additional follow-up and maximize the impact of the ReFRAME screening collection.
The website can be searched by structure or text string.
For Example searching for Malaria highlights a number of known therapeutic agents.
https://reframedb.org/#/search?query=malaria&type=string
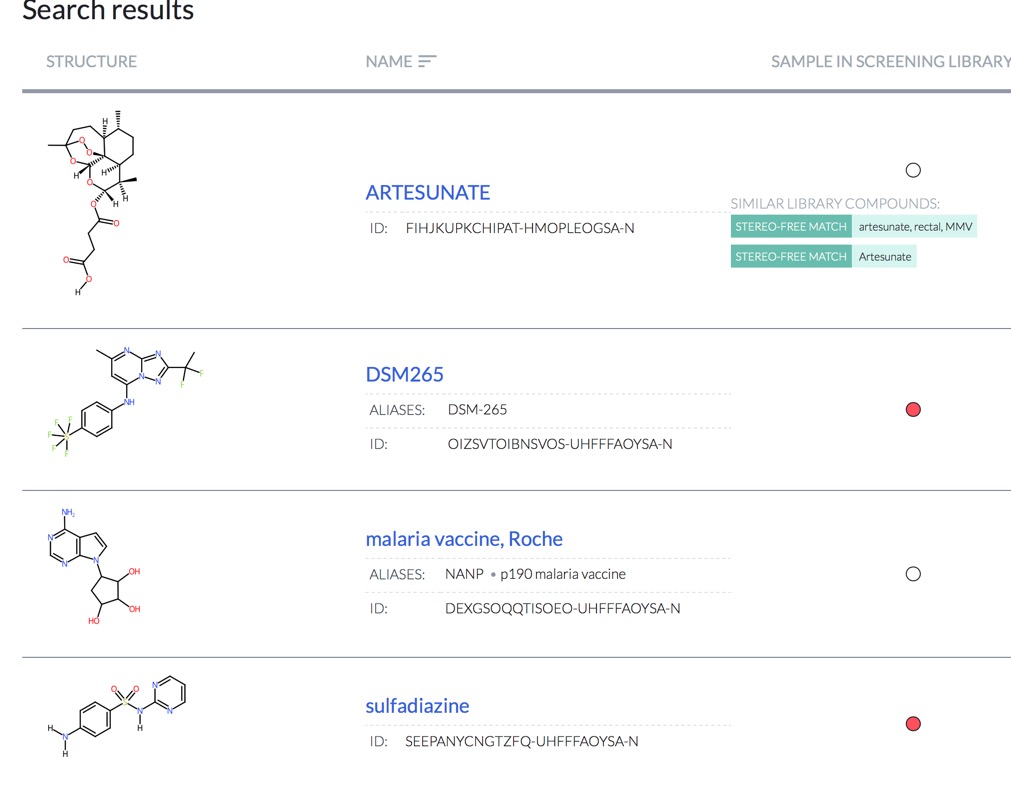
This looks like it will be an invaluable resource.
NAR Database Issue
The Nucleic Acid Research Database Issue is now available. Details of over 600 databases described in open access publications.
These databases cover a huge area of biological science, including:-
- Nucleic acid sequence, structure, and regulation
- Protein sequence and structure, motifs, and domains
- Metabolic and signalling pathways, enzymes
- Viruses, bacteria, protozoa and fungi
- Human genome, model organisms, comparative genomics
- Genomic variation, diseases, and drugs
- Plant databases
ChEMBL_23 released
ChEMBL_23 has been released, it was prepared on 1st May 2017 and contains:
- 2,101,843 compound records
- 1,735,442 compounds (of which 1,727,112 have mol files)
- 14,675,320 activities
- 1,302,147 assays
- 11,538 targets
- 67,722 source documents
Data can be downloaded from the ChEMBL ftp site: ftp://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/releases/chembl_23
Also includes
Deposited Data Sets
CO-ADD, The Community for Open Antimicrobial Drug Discovery, is a global open-access screening initiative launched in February 2015 to uncover significant and rich chemical diversity held outside of corporate screening collections. CO-ADD provides unencumbered free antimicrobial screening for any interested academic researcher. CO-ADD has been recognised as a novel approach in the fight against superbugs by the Wellcome Trust, who have provided funding through their Strategic Awards initiative. Open Source Malaria (OSM) is aimed at finding new medicines for malaria using open source drug discovery, where all data and ideas are freely shared, there are no barriers to participation, and no restriction by patents. The initial set of deposited data from the CO-ADD project consists of OSM compounds screened in CO-ADD assays (DOI = 10.6019/CHEMBL3832881).
Modelled on the Malaria Box, the MMV Pathogen Box contains 400 diverse, drug-like molecules active against neglected diseases of interest and is available free of charge (http://www.pathogenbox.org). The Pathogen Box compounds are supplied in 96-well plates, containing 10 uL of a 10mM dimethyl sulfoxide (DMSO) solution of each compound. Upon request, researchers around the world will receive a Pathogen Box of molecules to help catalyse neglected disease drug discovery. In return, researchers are asked to share any data generated in the public domain within 2 years, creating an open and collaborative forum for neglected diseases drug research. The initial set of assay data provided by MMV has now been included in ChEMBL (DOI = 10.6019/CHEMBL3832761).
SkinSensDB added to drug discovery resources
I've added SkinSensDB to the Drug Discovery Resources page covering Chemistry and Biology Databases.
Skin sensitization is an important toxicological endpoint for chemical hazard determination and safety assessment….SkinSensDB has been constructed by curating data from published AOP-related assays. In addition to providing datasets for developing computational models, SkinSensDB is equipped with browsing and search tools which enable the assessment of new compounds for their skin sensitization potentials based on data from structurally similar compounds.
SkinSensDB: a curated database for skin sensitization assays DOI.
Nucleic Acid Research Database Issue
I've just been browsing through the NAR Database issue whilst the the historical focus was on molecular biology resources the current issue contains over 500 resources that would be of interest to drug discovery. In addition to well known databases like ChEMBL, PubChem BioAssay and RCSB there are many others. A few that caught my eye:
TransportersDB Database of membrane transporters
SureChEMBL Annotated patent database
canSAR Cancer research and drug discovery knowledgebase
iPPI-DB Modulators of protein-protein interactions
Withdrawn Withdrawn drugs
Open Targets platform for therapeutic target identification and validation
ChEMBL 22 Released
ChEMBL 22 has been released. ChEMBL is a database of bioactive drug-like small molecules, it contains 2-D structures, calculated properties (e.g. logP, Molecular Weight, Lipinski Parameters, etc.) and abstracted bioactivities (e.g. binding constants, pharmacology and ADMET data).
This version of the database, prepared on 8th August 2016 contains:
- 2,043,051 compound records
- 1,686,695 compounds (of which 1,678,393 have mol files)
- 14,371,219 activities
- 1,246,132 assays
- 11,224 targets
- 65,213 documents
There is more information in the ChEMBL blog post
ChEMBL 18 released
ChEMBL_18 has just been released.
It can be downloaded from the ChEMBL FTP site, and there are more details on the ChEMBL blog
- 1,566,466 compound records
- 1,359,508 compounds (of which 1,352,681 have mol files)
- 12,419,715 activities
- 1,042,374 assays
- 9,414 targets
- 53,298 documents
They now include epigenetic targets, and several new web services giving drug approvals and mechanisms.
SureChEMBL
I just saw a note that the ChEMBL group that they are taking over the running of the SureChem system from Digital Science. This means that ChEMBL will be collating the chemical patent literature.
More details from the announcement
For those of you that are already SureChem users you will be familiar with the functionality and how it works; but for those that weren't SureChEMBL takes feeds of full text patents, identifies chemical objects from either the in-line text or from images and adds 2-D chemical structures. This is then loaded into a database and is searchable by chemical structure, so you can do substructure, similarity searching and so forth - all the good things you'd expect from a chemical database. This chemical search functionality is unavailable from the public, published patent documents, and is really essential for anyone seriously using the patent literature. Oh, and the system does this live, so as patents are published, they are processed and added to the system - the delay between publication and structures being available in SureChEMBL is about a day when converted from text, and a few days when converted from image sources
RSC aquires The Merck Index
The Merck Index, is to join the highly acclaimed publishing portfolio of the Royal Society of Chemistry.
The RSC already plans significant development of The Merck Index online, and will continue to develop and update.
I wonder if this will be integrated into ChemSpider? When you also consider that the RSC is taking over the Chemical Database Service it is clear that the RSC is moving to increase it’s support of online chemistry.
Following a tendering exercise earlier this year the EPSRC will be renewing the Chemical Database Facility, with the Royal Society of Chemistry being the preferred bidder. Contract negotiations are being pursued and should be in place by the end of this year. Details of the renewed service will be made known in due course and the RSC will be working with Daresbury to ensure a seamless transfer to the new system.
Zinc databases Updated
I see the Zinc databases have been updated (ZINC, is a free database of commercially-available compounds for virtual screening)
Lead-like, 5.7M, today Shards, 55K, Oct 15 Frag-like, 636K, Oct 15 Leads-now, 2M, Oct 15 Frags-now, 440K, Oct 12
There are more databases here
Updated Pages
I’ve updated the page of online databases to include databases that contain biological information that might be useful.
I’ve also added a link to a series of presentations and posters available from Sirius on the physicochemical properties page.
ChEMBL 13
ChEMBL 13 has been released.
This release includes updates to the manually extracted Medicinal Chemistry literature, updates to OrangeBook drug approvals and a update from PubChem BioAssay. This release also contains data sets related to screening against human African Trypanosomiasis and Chagas disease. Both data sets have been deposited by the Drugs for Neglected Diseases Initiative (DNDi).
This latest version of the ChEMBL database contains:
- 1,304,115 compound records
- 1,143,682 distinct compounds
- 617,681 assays
- 6,933,068 bioactivities
- 8,845 targets
- 44,68 documents
- 8 data sources
The data can be downloaded from the ChEMBL website.
Chemical Databases
Updated the Chemical Databases page, added MMsINCdatabase a free web-oriented database of commercially-available compounds for virtual screening and chemoinformatic applications. MMsINC contains over 4 million non-redundant chemical compounds in 3D formats.
ChemSpider Search
If like me you regularly come across drugs mentioned on web pages that you are unfamiliar with then this Safari Extension will be of interest. If a page contains a drug name (this page describes AOX1 substrates), select the name and right click (or control click) and an option appears to search for the highlighted drug on ChemSpider.
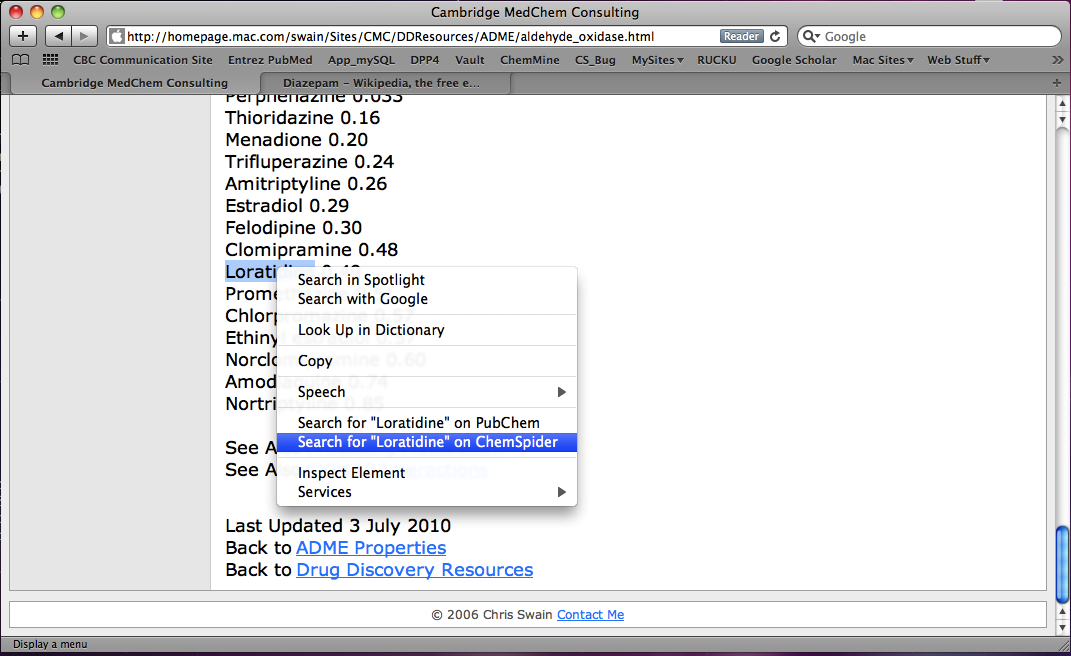
Click on Search for “Loratidine” on ChemSpider option and the structure appears in a small window.
The new version of the extension has a few extra options, the small window that pops up containing the structure now has a number of additional options highlighted below, if you click on the “3D” button the display changes to a 3D rendering using the Java applet JMOL. If you now click on the “Zoom” button.
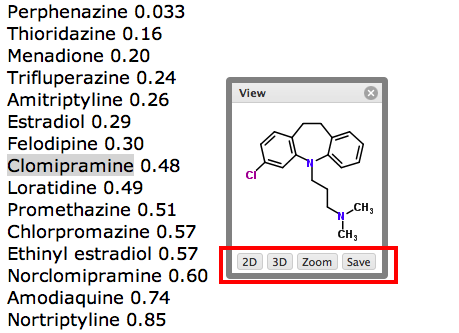
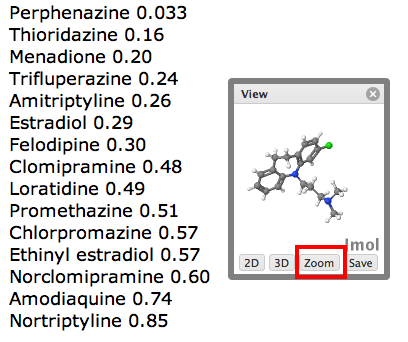
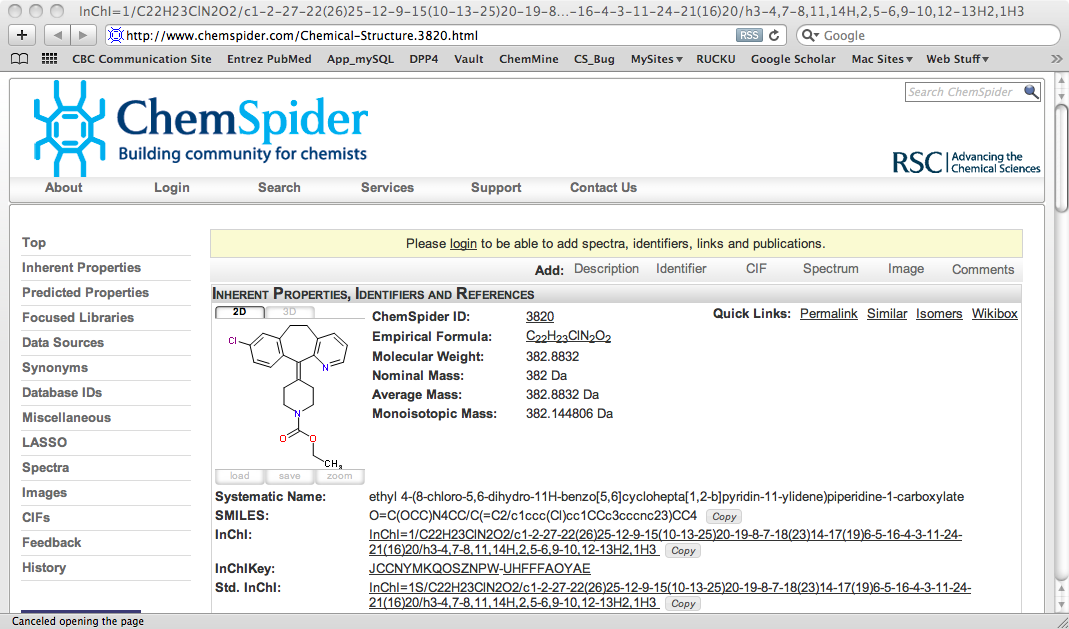
If you then click on the “view” option a new page opens showing the full details in ChemSpider.
Updated Pages
Updated the Chemical Databases and Physicochemical Properties pages of the Drug Discovery Resources.