Annual Site Review
As 2015 ends I'd like to take the chance to wish you all a Happy New Year and hope for great success in your drug discovery endeavours.
The website increases in popularity with 93,000 page views in 2015 an increase of 24% over last year. Nearly 25% of the visitors come back on multiple occasions which I hope means people are finding the content useful.
Nine of the top ten most popular pages were from the Drug Discovery Resources Pages which I am delighted to see, since it suggests that the work entailed in putting the resources together is worthwhile.
The most viewed pages were
- Distribution and Plasma Protein Binding
- Calculating Physicochemical Properties
- Molecular Interactions
- Lipophilicity
- Formulation
- Bioisosteres
- Fragment based screening
- Aspartic Acid Protease Inhibitors
- Serine Protease Inhibitors
As might be expected the Books page only seems popular coming up to Christmas ;-)
The visitors come from over 100 different countries with US and UK topping the list. Whilst desktop systems predominate nearly 20% now access the site from a mobile device.
Insects in Drug Discovery
The company N2MO offers the use of insects as model organisms. They can be used for ADME screening in particular brain penetration studies.
The Grasshopper: A Novel Model for Assessing Vertebrate Brain UptakeOlga Andersson, Steen Honoré Hansen, Karin Hellman, Line Rørbæk Olsen, Gunnar Andersson, Lassina Badolo, Niels Svenstrup, and Peter Aadal Nielsen EntomoPharm R&D, Medicon Village, Lund, Sweden (O.A., K.H., G.A., P.A.N.); Department of Pharmacy, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark (S.H.H., L.R.O.); and Division of Discovery Chemistry and Drug Metabolism and Pharmacokinetics, H. Lundbeck A/S, Copenhagen, Denmark (L.B., N.S.) Received April 10, 2013; accepted May 10, 2013
ABSTRACT The aim of the present study was to develop a blood-brain barrier (BBB) permeability model that is applicable in the drug discovery phase. The BBB ensures proper neural function, but it restricts many drugs from entering the brain, and this complicates the development of new drugs against central nervous system diseases. Many in vitro models have been developed to predict BBB permeability, but the permeability characteristics of the human BBB are notoriously complex and hard to predict.
Consequently, one single suitable BBB permeability screening model, which is generally applicable in the early drug discovery phase, does not yet exist. A new refined ex vivo insect-based BBB screening model that uses an intact, viable whole brain under controlled in vitro-like exposure conditions is presented.
This model uses intact brains from desert locusts, which are placed in a well containing the compound solubilized in an insect buffer. After a limited time, the brain is removed and the compound concentration in the brain is measured by conventional liquid chromatography-mass spectrometry. The data presented here include 25 known drugs, and the data show that the ex vivo insect model can be used to measure the brain uptake over the hemolymph-brain barrier of drugs and that the brain uptake shows linear correlation with in situ perfusion data obtainedinvertebrates.Moreover,this study shows that the insect ex vivo model is able to identify P-glycoprotein (Pgp) substrates, and the model allows differentiation between low-permeability compounds and compounds that are Pgp substrates.
Target Validation
There has been much discussion about the attrition of drugs in development due to lack of efficacy in man and this in part can be due to poor target validation. That is proof that modulation of the identified target in a model system has the desired impact on biological activity and can be linked to therapeutic utility.
This is an absolutely critical step, almost everything else can be fixed.
For this reason two new resources seem particularly valuable.
The Centre for Therapeutic Target Validation platform (https://www.targetvalidation.org) brings together information on the relationships between potential drug targets and diseases. The core concept is to identify evidence of an association between a target and disease from various data types.
A target can be a protein, protein complex or RNA molecule, but we integrate evidence through the gene that codes for the target. In the same way, we describe diseases through a structure of relationships called the Experimental Factor Ontology (EFO) that allows us to bring together evidence across different but related diseases.The platform supports workflows starting from either a target or disease and presents the evidence for target – disease associations in a number of ways through association and evidence pages.
DisGeNET(http://www.disgenet.org/web/DisGeNET/menu/home) is a discovery platform integrating information on gene-disease associations (GDAs) from several public data sources and the literature doi.
The current version contains (DisGeNET v3.0) contains 429111 associations, between 17181 genes and 14619 diseases, disorders and clinical or abnormal human phenotypes.
Drug Discovery Resources Update
I've updated the hit identification section of the Drug Discovery Resources. In particular I've added to the high-throughput screening analysis including more information on PAINS (Pan Assay Interference Compounds) first described by Baell et al DOI and subsequently summarised in an excellent Nature comment.
Academic researchers, drawn into drug discovery without appropriate guidance, are doing muddled science. When biologists identify a protein that contributes to disease, they hunt for chemical compounds that bind to the protein and affect its activity. A typical assay screens many thousands of chemicals. ‘Hits’ become tools for studying the disease, as well as starting points in the hunt for treatments.
These molecules — pan-assay interference compounds, or PAINS — have defined structures, covering several classes of compound. But biologists and inexperienced chemists rarely recognize them. Instead, such compounds are reported as having promising activity against a wide variety of proteins. Time and research money are consequently wasted in attempts to optimize the activity of these compounds. Chemists make multiple analogues of apparent hits hoping to improve the ‘fit’ between protein and compound. Meanwhile, true hits with real potential are neglected.
Also added a page on Aggregators. Promiscuous inhibition caused by small molecule aggregation is a major source of false positive results in high-throughput screening. To mitigate this, use of a nonionic detergent such as Triton X-100 or Tween-80 has been studied, which can disrupt aggregates, and is now common in screening campaigns DOI.
The Handbook of Medicinal Chemistry
The Handbook of Medicinal Chemistry is a new book providing insight and advice for medicinal chemists.
Drug discovery is a constantly developing and expanding area of research. Developed to provide a comprehensive guide, the Handbook of Medicinal Chemistry covers the past, present and future of the entire drug development process. Highlighting the recent successes and failures in drug discovery, the book helps readers to understand the factors governing modern drug discovery from the initial concept through to a marketed medicine. With chapters covering a wide range of topics from drug discovery processes and optimization, development of synthetic routes, pharmaceutical properties and computational biology, the handbook aims to enable medicinal chemists to apply their academic understanding to every aspect of drug discovery. Each chapter includes expert advice to not only provide a rigorous understanding of the principles being discussed, but to provide useful hints and tips gained from within the pharmaceutical industry. This expertise, combined with project case studies, highlighting and discussing all areas of successful projects, make this an essential handbook for all those involved in pharmaceutical development.
A free app has also been created in collaboration with the editors of the book. The Medicinal Chemistry Toolkit provides a suite of resources to support the day to day work of a medicinal chemist
Do Privileged PPI Scaffolds Exist
I've been working with the BioFocus group at Chesterford Park (now part of Charles River) thinking about ligands for Protein Protein Interactions, some of the work was described on a poster at the 18th Cambridge Medicinal Chemistry Meeting held in Cambridge in September this year. The poster is now available online http://www.criver.com/files/pdfs/nonsource/do-privileged-ppi-scaffolds-exist.aspx
Protein-protein interactions (PPIs) are ubiquitous in cellular biochemistry; however they are often difficult drug targets to interrogate due to their unique molecular topologies. A consequence is that low hit rates are frequently observed in PPI HTS campaigns and there remains an unmet need for innovative small molecule PPI inhibitors (SMPPIIs). The term "privileged scaffold" was coined in 1988 when core structures were found to bind to more than one receptor with high affinity. This led us to pose the question: “Do privileged PPI scaffolds exist?”
A brilliant group of scientists to work with, many stimulating discussions in a very important area.
CRUK Grand Challenges
Cancer Research UK has a series of 7 grand challenges detailed on its website
Challenge 1: Develop vaccines to prevent non-viral cancers
Challenge 2: Eradicate EBV-induced cancers from the world
Challenge 3: Identify new targets for cancer prevention by understanding how unusual patterns of mutation are induced by different cancer-causing events
Challenge 4: Distinguish between lethal cancers that need treating, and non-lethal cancers that don’t
Challenge 5: Map the molecular and cellular tumour microenvironment in order to define new targets for therapy and prognosis
Challenge 7: Deliver biologically active macromolecules to any and all cells in the body to effectively treat cancer
Well worth reading about and there is also the opportunity to suggest your own challenge.
Setting up Cheminformatics Support for the Open Source Malaria Project
Here are the slides for a talk I gave at the Cambridge Cheminformatics Network Meeting in August. Setting up Cheminformatics Support for the Open Source Malaria Project
Latest Publication
Bifunctional crosslinking ligands for transthyretin
P. Patrizia Mangione, Stéphanie Deroo, Stephan Ellmerich, Vittorio Bellotti, Simon Kolstoe, Stephen P. Wood, Carol V. Robinson, Martin D. Smith, Glenys A. Tennent, Robert J. Broadbridge, Claire E. Council, Joanne R. Thurston, Victoria A. Steadman, Antonio K. Vong, Christopher J. Swain, Mark B. Pepys, Graham W. Taylor
Wild-type and variant forms of transthyretin (TTR), a normal plasma protein, are amyloidogenic and can be deposited in the tissues as amyloid fibrils causing acquired and hereditary systemic TTR amyloidosis, a debilitating and usually fatal disease. Reduction in the abundance of amyloid fibril precursor proteins arrests amyloid deposition and halts disease progression in all forms of amyloidosis including TTR type. Our previous demonstration that circulating serum amyloid P component (SAP) is efficiently depleted by administration of a specific small molecule ligand compound, that non-covalently crosslinks pairs of SAP molecules, suggested that TTR may be also amenable to this approach. We first confirmed that chemically crosslinked human TTR is rapidly cleared from the circulation in mice. In order to crosslink pairs of TTR molecules, promote their accelerated clearance and thus therapeutically deplete plasma TTR, we prepared a range of bivalent specific ligands for the thyroxine binding sites of TTR. Non-covalently bound human TTR–ligand complexes were formed that were stable in vitro and in vivo, but they were not cleared from the plasma of mice in vivo more rapidly than native uncomplexed TTR. Therapeutic depletion of circulating TTR will require additional mechanisms.
Hepatotoxicity
I've expended the preclinical toxicity section to include a page on hepatotoxicity. This gives some details of the common assays and markers used to evaluate the potential hepatotoxicity.
An Aggregation Advisor for Ligand Discovery
Aggregation is a regular concern when evaluating potential hits from screening and a recent paper "An Aggregation Advisor for Ligand Discovery" DOI attempts to provide an insight into this phenomenon, in addition they provide a useful web-based tool http://advisor.bkslab.org that provides a free service to advise whether molecules may aggregate under biological assay conditions.
Time Dependent Inhibition
I've just updated the Drug Discovery Resources page on CYP Interactions, included a section on Time Dependent Inhibition (TDI).
ResearchKit
An article on BuzzFeed suggests that Pharma companies are investigating the use of ResearchKit in clinical trial studies.
ResearchKit is an open source framework introduced by Apple that allows researchers and developers to create powerful apps for medical research. Easily create visual consent flows, real-time dynamic active tasks, and surveys using a variety of customizable modules that you can build upon and share with the community. And since ResearchKit works seamlessly with HealthKit, researchers can access even more relevant data for their studies — like daily step counts, calorie use, and heart rate
GlaxoSmithKline apparently is currently working on integrating (ResearchKit) into clinical trials and planning to start in coming months, whilst Purdue Pharma are in the early stages of exploring whether Apple’s new tool for research data collection can be used as part of its own drug R&D efforts.
So far, ResearchKit apps are being led by academic medical centers like the University of California, San Francisco, and nonprofits like Sage Bionetworks and the Michael J. Fox Foundation for Parkinson’s Research. LifeMap Solutions, a company that develops mobile health apps, helped create the asthma app in partnership with the Icahn School of Medicine at Mount Sinai. The first ResearchKit apps signed up more than 75,000 participants in just the first few months
Antibiotic Screening
A little while ago I mentioned The Community for Open Antimicrobial Drug Discovery effort to provide free compound screening against a variety of infective agents. I now have a few more details of what you might be able to access for a 1mg sample.
Primary Screening:-
Test against key ESKAPE pathogens, E. coli,
K. pneumoniae, A. baumannii, P. aeruginosa,
S. aureus (MRSA), as well as the fungi C. neoformans and C. albicans, at a single concentration.
Hit Confirmation:-
Confirm activity with minimum inhibitory concentration and counterscreen for cytotoxicity and membrane interaction.
Hit Validation:-
Test the positive hit against a broader panel of microbes and evaluate the basic drug qualities of actives.
CO-ADD will screen your compounds for free and make no claim to IP
Metrabase
p>The Metabolism and Transport Database (Metrabase) is a cheminformatics and bioinformatics resource that contains curated data related to human small molecule metabolism and transport, Journal of Cheminformatics 2015, 7:31 DOI. Currently it includes interaction data on 20 transporters, 3438 molecules and 11649 interaction records manually abstracted from 1211 literature references and supplemented with data from other resources as shown in the image below taken from the original publication.
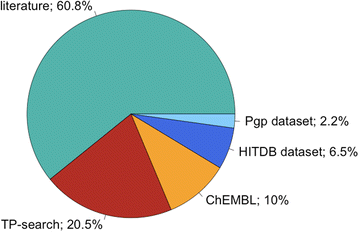
I've added this and more details to the Transporters page of the Drug Discovery Resources
Grant Funding Research
I've just updated the page listing possible sources of grant funding for drug discovery research. In particular I've extended the listing of disease specific resources, these may be particularly useful for rare or neglected diseases.
Paracetamol Challenge
Sadly it appears that the latest craze to sweep social media is the Paracetamol Challenge in which people (usually children) are encouraged to consume large amounts of the over the counter analgesic paracetamol (called acetaminophen in North America).
One of the particularly insidious features of paracetamol toxicity is that individuals may display little or no symptoms in the first 24h, it is only later when increasing liver damage has occurred do the more serious symptoms become apparent.
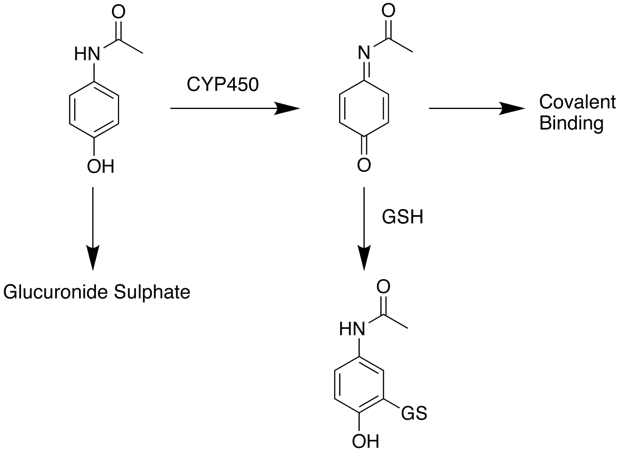
What is the mechanism of Paracetamol Toxicity
At normal therapeutic doses paracetamol the main route for clearance is by Phase II processes including conjugation to form the glucuronide or sulphate followed by renal clearance. At higher doses however, paracetamol is oxidised by CYP450 enzymes in the liver to a highly reactive intermediary metabolite, N-acetyl-p-benzoquinoneimine (NAPQI). NAPQI can be detoxified by conjugation with glutathione (GSH) to form cysteine and mercapturic acid conjugates however this pathway has limited capacity and once supplies of glutathione are exhausted if NAPQI remains it can react covalently with biomolecules resulting in widespread hepatocyte damage and death, leading to acute hepatic necrosis. Without treatment this progresses to irreversible liver, kidney failure followed by multiple organ failures.
Treatment
If caught within an hour of ingestion gastric lavage may be used to remove any drug that has not yet been absorbed, at later stages N-acetylcysteine can be administered. This works to reduce paracetamol toxicity by replenishing body stores of glutathione (GSH). However N-acetylcysteine needs to be administered before liver damage has occurred, if given more than 8 hours after ingestion of paracetamol it's effectiveness is reduced. If patients develop hepatic failure or who are otherwise expected to die from liver failure, the mainstay of treatment is liver transplantation.
Lack of reproducibility with antibodies
A slightly worrying article in Nature, Reproducibility crisis: Blame it on the antibodies.
The lack of reproducibility of published data on potential drug targets has been highlighted on several occasions DOI and it has been suggested that this is a major factor in the failure rate for phase 2 clinical trials DOI.
In almost two-thirds of the projects, there were inconsistencies between published data and in-house data that either considerably prolonged the duration of the target validation process or, in most cases, resulted in termination of the projects.
Antibodies have rapidly become a key tool in understanding and identifying new drug targets and potentially used as biomarkers to identify patients. However it is clear that many of the 2 million commercially available antibodies need to be checked rigorously, with some scientists claiming more than half are unreliable.
In 2011, an evaluation4 of 246 antibodies used in epigenetic studies found that one-quarter failed tests for specificity, meaning that they often bound to more than one target. Four antibodies were perfectly specific — but to the wrong target.
Caveat emptor.
Medicinal Chemistry Toolkit app
A review of the Medicinal Chemistry Toolkit app for iOS
The role of solvent in ligand binding
p>The role of water in ligand binding is often ignored and I thought it might be useful to add information to the drug discovery resources section. In particular :-
Expanding the page on molecular interactions
and adding a page on Solvation and Desolvation
If you have time to have a look, any comments or suggestions would be very welcome.
European Lead Factory
p>The European Lead Factory has just announced that an additional 50,000 new compounds have been added to their screening collection. This brings the collection up to 350,000 compounds and sets them well on the way to their 500,000 target.
I've been involved in a couple of projects that have made use of this high-throughput screening facility and I've been impressed with the quality and diversity of the hits generated.
The European Lead Factory was established to promote the discovery of novel lead compounds, suitable for subsequent optimization either to drug candidates or to high‐quality pharmacological tools for the experimental validation of targets.
If you have a target you want to screen against you can submit a proposal online. For an academic or small company this is an interesting way to identify novel starting points for a medicinal chemistry program.
Free Compound Screening for antimicrobial activity
The concerns about antibiotic resistance are well known and indeed have made headlines in the mainstream press. Here is a chance to help find the next generation of antibiotics.
Do you have interesting compounds sitting on the shelf? Perhaps you would be interested in having them screened for antibiotic activity for free?
The Community for Open Antimicrobial Drug Discovery would like to hear from you, their goal is to screen compounds from academic research groups from anywhere in the world for free.
The requirements are pretty minimal
We ask for 1-2 mg of pure compound which will be used for primary screening, hit confirmation, and if active will be used for a broader antimicrobial screening, cytotoxicity and a check for its purity. We require all compounds to be soluble in water or DMSO and to be shipped as dry material in appropriate containers, such as 1-2 mL Eppendorf tubes. For larger collections we can arrange plates or tube-racks.
In the primary screen they test against against key ESKAPE pathogens, E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, S. aureus (MRSA), as well as the fungi C. neoformans and C. albicans. The ‘ESKAPE’ pathogens that are responsible for two-thirds of all health care-associated infections and resistant strains of these bacteria represent the greatest unmet need in antibacterial drug development.
New Compound Sets Identified from High Throughput Phenotypic Screening Against Three Kinetoplastid Parasites: An Open Resource
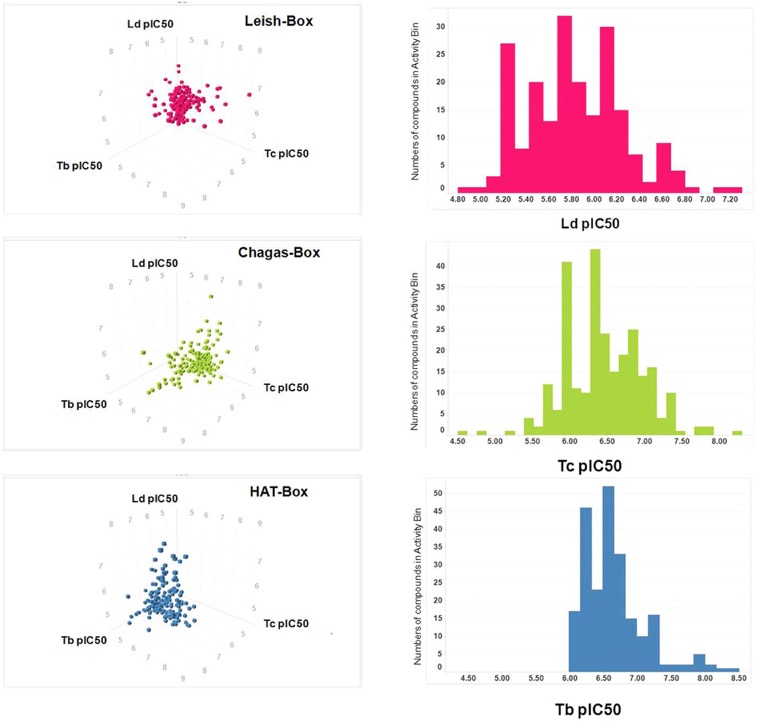
The GSK high-throughput screening group at Tres Cantos and collaborators have just published DOI the results of whole-cell phenotypic screens against the three kinetoplastids most relevant to human disease, i.e. Leishmania donovani, Trypanosoma cruzi and Trypanosoma brucei. Three anti-kinetoplastid chemical boxes of ~200 compounds each were assembled. Functional analyses of these compounds suggest a wide array of potential modes of action against kinetoplastid kinases, proteases and cytochromes as well as potential host-pathogen targets. The compound sets are provided as an open resource for the scientific community.
Caveat emptor
A paper entitled Promiscuous 2-Aminothiazoles (PrATs): A Frequent Hitting Scaffold appeared in J Med Chem recently DOI, in which they describe the promiscuous nature of 2-aminothiazoles in screens.
Exemplified by 4-phenylthiazol-2-amine being identified as a hit in 14/14 screens against a diverse range of protein targets, suggesting that this scaffold is a poor starting point for fragment-based drug discovery
I thought I'd check how often this substructure appears in the published fragments database, indeed currently 43 of the 903 published fragments contain this substructure. Further investigation identifies a total of 63 amino-substituted 5-membered heterocycles, and there are 167 fragments in which there is an amino group on an aromatic ring (mainly heterocycles).
It should also be noted however that there are 64 structures in the DrugBank database that also contain a 2-aminothiazole, so whilst promiscuous they can be developed into drugs.
So whether they are privileged structures or troublesome promiscuous hits is probably in the eye of the beholder, caveat emptor.
Annual Site Review
At the end of each year I take the opportunity to look at the website analytics to see what parts of the website are the most popular. Overall there was a 15% increase in the number of page views up to 75,000. Average time on a page was 2 mins suggesting the content is engaging with the viewers.
Nine of the top ten most popular pages were from the Drug Discovery Resources Pages which I am delighted to see, since it suggests that the work entailed in putting the resources together is worthwhile.
The most viewed pages were
More on PAINS
I often get asked to help with the analysis of high-throughput screening results and one of the first filters I run as part of the hit identification is to flag for PAINS (Pan Assay Interference Compounds) first described by Baell et al DOI and subsequently summarised in an excellent Nature comment.
Academic researchers, drawn into drug discovery without appropriate guidance, are doing muddled science. When biologists identify a protein that contributes to disease, they hunt for chemical compounds that bind to the protein and affect its activity. A typical assay screens many thousands of chemicals. ‘Hits’ become tools for studying the disease, as well as starting points in the hunt for treatments.
These molecules — pan-assay interference compounds, or PAINS — have defined structures, covering several classes of compound. But biologists and inexperienced chemists rarely recognize them. Instead, such compounds are reported as having promising activity against a wide variety of proteins. Time and research money are consequently wasted in attempts to optimize the activity of these compounds. Chemists make multiple analogues of apparent hits hoping to improve the ‘fit’ between protein and compound. Meanwhile, true hits with real potential are neglected.
In the supplementary information they provided the corresponding filters in Sybyl Line Notation (SLN) format, however they have also been converted to SMARTS format and incorporated in sieve file for use in filtering compound collections. If you are a Vortex user then there is also a Vortex script available, filters are also available for Knime and now it is even available on mobile devices with MolPrime+.
It is probably not until you have been involved in multiple small molecule screens that you appreciate the number of ways that false positives can occur and just how much valuable time and resources can be wasted following them up. Indeed it may be for the more difficult targets the majority of hits seen may be false positives. Flagging PAINS is now such a well developed tool that it would be fool hardy not to include it.