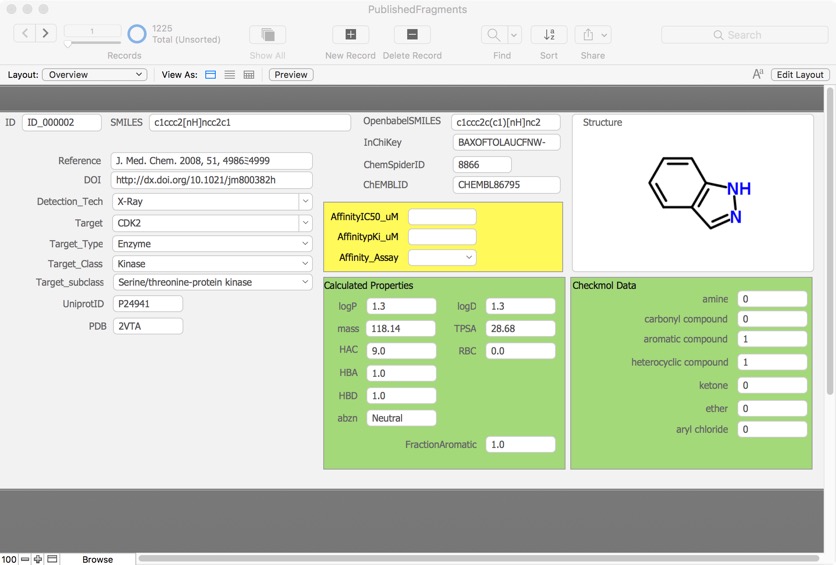
Deconstruction of a screening hit
One strategy to investigate screening hits is to simplify the structure to identify the minimum pharmacophore . Whilst lower in affinity the result will have lower molecular weight and LogP. Indeed the molecule may now occupy "fragment space". When I presented this at the recent Fragments meeting a member of the audience coined the phrase "Deconstruction of a screening hit" and several people used the phrase subsequently so perhaps it will used in the future.
Details are here Deconstruction of a screening hit.
9th Fragment-based Drug Discovery Meeting
I'll be heading over to the 9th Fragment-based Drug Discovery Meeting https://www.rscbmcs.org/events/fragments24/ later today. This event is one of the high points in the Drug Discovery calendar. I'm sure there will be plenty to add to the Fragment-Based Screening section on the Drug Discovery Resources Website.
The aim of the 9th RSC-BMCS Fragment-based Drug Discovery meeting will be to continue the focus on case studies in Fragment-based Drug Discovery that have delivered compounds to late stage medicinal chemistry, preclinical or clinical programmes. The Fragment series was started in 2007 and continues with this theme in having over three-quarters of the presentations focused on case studies. This will be complemented by technology progress in high concentration, NMR, SPR and X-ray screening.
Updated Hit identification page
The Hit Identification page to include information from a great analysis published recently. An Analysis of Successful Hit-to-Clinical Candidate Pairs?" DOI.
Ultra large Chemical Libraries
In a recent blog post Derek Lowe talked about "Virtual Screening Versus the Numbers" https://www.science.org/content/blog-post/virtual-screening-versus-numbers highlighting some of the issues around ultra large chemical libraries.
It seems quite timely that RSC CICAG is organising a meeting on Ultra Large Chemical libraries 10 August 2022 10:00-17:00, Burlington House, London, United Kingdom.
A decade ago a chemical library of a million compounds was considered large but over the last few years there has been a period of continuous growth in the size of both physical and virtual chemical libraries. As the libraries have grown the conventional search technologies have become unsustainable and new technologies are needed. This meeting will look at the challenges and solutions used to design, create, compare and search these ultra-large chemical libraries.
There are more details and registration here https://www.rsc.org/events/detail/73675/ultra-large-chemical-libraries.
It is now open for abstract submission (oral due by May 1st, posters June 2nd).
Registration fees
Delegate member early £95
Delegate non-member early £115
Delegate member std £120
Delegate non-member std £145
Student member early £65
Student non-member early £85
Student member std £90
Student non-member std £110
Find out more about the European Lead Factory
On Tuesday 30 November, members of the European Lead Factory (ELF) will participate in a webinar organised by the Young Scientists Network (YSN) of the European Federation of Medicinal Chemistry and Chemical Biology (EFMC).
The topic of the session is “Biological Testing of Hit Compounds”. Dr Vera Nies, Programme Manager at Lygature (the coordinating partner of the ELF) will give an introduction to the European Lead Factory. Her talk will be followed by a presentation entitled "Best practices in High Throughput Screening: ELF as an example", by Dr Steven van Helden of Pivot Park Screening Centre. The last ELF member presentation will be given by Dr Phil Jones of BioAscent, who will talk about the “Approaches Towards PAINSless Lead Generation”.
More details and registration here https://www.europeanleadfactory.eu/newsroom/elf-participate-efmc-ysn-webinar
ELF programs available for partnering
The European Lead Factory (ELF) is a collaborative public-private partnership aiming to deliver innovative drug discovery starting points. The ELFgives free access to up to 550,000 novel compounds, a unique industry-standard uHTS platform, and much more.
The success of the ELF approach has been widely acknowledged and the output has shown to be of high quality, worth following up and investment-ready. Funding for further development has been secured for several programmes, and by March 2018 two programmes have led to partnering deals being closed between the Programme Owner and an established pharmaceutical company.
A number of projects are now available for partnering.
The available projects are here.
European Lead Factory call for proposals
The closing date for submissions to the European Lead Factory is Friday 28th May 2021.
Following the UK’s exit from the European Union on 31 January, we are pleased to confirm that the European Lead Factory still welcomes screening proposals from UK researchers.
This is a fantastic opportunity for researchers to get access to a top class high-throughput screening platform.
On this page, you can find the application forms, legal documents and all other relevant information required for your proposal submission.
The screening deck consists of around 535,000 compounds consisting of about 300,000 compounds from the original pharmaceutical companies, along with almost 190,000 compounds from custom design and synthesis. Furthermore, Grünenthal and Institut de Recherche Servier have provided around 50,000 new compounds not previously available to the European Lead Factory. Compounds from partners were selected for inclusion in the library based on several important quality criteria, as well as on the prerequisite that they are unique and not commercially available. Efforts were also taken to increase diversity, resulting in a top-quality compound library.
March webinar: High Content Screening at the European Lead Factory
The next European Lead Factory webinar will take place on Thursday 11 March from 11:00-12:00 (CET). In this webinar, we will focus on High Content Screening with a presentation by Professor Jason Swedlow, head of the National Phenotypic Screening Centre at the University of Dundee.
More details and registration here https://www.europeanleadfactory.eu/node/375
The single most important factor determining the likelihood of success of a project is the quality of the starting lead
Have you identified a disease mechanism that should be explored in a drug discovery project? Then you may have also considered developing a screening assay to find new chemical starting points that could serve as candidates for drug development.
In the next European Lead Factory webinar, taking place on Thursday 28 January, Dr Saman Honarnejad, Director of Drug Discovery at Pivot Park Screening Centre, will give a presentation on guidelines for HTS assay development, with an emphasis on methods to design, evaluate and improve conditions for biochemical and cellular assay formats.
The event will take place via Microsoft Teams on 28 January 2021 from 11:00-12:00 (CET). Discussions with the audience will form a key part of the webinar and time will be made available for questions and answers.
Registration is free using this link https://events.europeanleadfactory.eu/elf/elf-webinar/.
The European Lead Factory welcomes drug targets in all therapeutic areas. Interested? Submit your screening proposal now. The next deadline to apply will be in January 2021. Full details are on the ELF website.
I've been involved in a number of screening campaigns and I can confirm this is a high quality screening deck.
Still time to submit proposal to European Lead Factory
The closure date for the latest round of proposals for screening at the European Lead Factory is January 2021
The European Lead Factory welcomes drug targets in all therapeutic areas. Interested? Submit your screening proposal now. The next deadline to apply will be in January 2021.
Full details are on the ELF website.
I've been involved in a number of screening campaigns and I can confirm this is a high quality screening deck.
Proposals for European Lead Factory
The closure date for the latest round of proposals for screening at the European Lead Factory is January 2021
The European Lead Factory welcomes drug targets in all therapeutic areas. Interested? Submit your screening proposal now. The next deadline to apply will be in January 2021.
Also note
Following the UK’s exit from the European Union on 31 January, we are pleased to confirm that the European Lead Factory still welcomes screening proposals from UK researchers.
You can read more about the screening facility here https://www.europeanleadfactory.eu/node/353.
The details of how to submit are here https://www.europeanleadfactory.eu/how-submit/drug-target-assays/how-it-works.
I've written about the ELF here.

European Lead Factory Q&A
Interested in accessing a high quality high-throughput screening platform? Here is a chance to find out more about the European Lead Factory.
More details are here
The European Lead Factory (ELF) is a collaborative public-private partnership aiming to deliver novel lead molecules for drug discovery programs.
I've previously written about the ELF here.
Fragment based screening pages updated
I spent some time over the Christmas break updating the Drug Discovery Resources pages on Fragment-Based screening, adding new vendors and updating the physicochemical profiles. I've also added some discussion on the elaboration/optimisation of fragments.
The pages are
Fragment-Based Screening
Building a Fragment Collection
Available Fragment Collections
Profiles of Fragment Collections
Fragment-Based Screening Published Hits
The published fragments contains details of fragments that have been reported as hits in the literature, this database now has over 1500 entries culled from over 310 publications directed at nearly 220 different molecular targets using 26 different detection technologies.
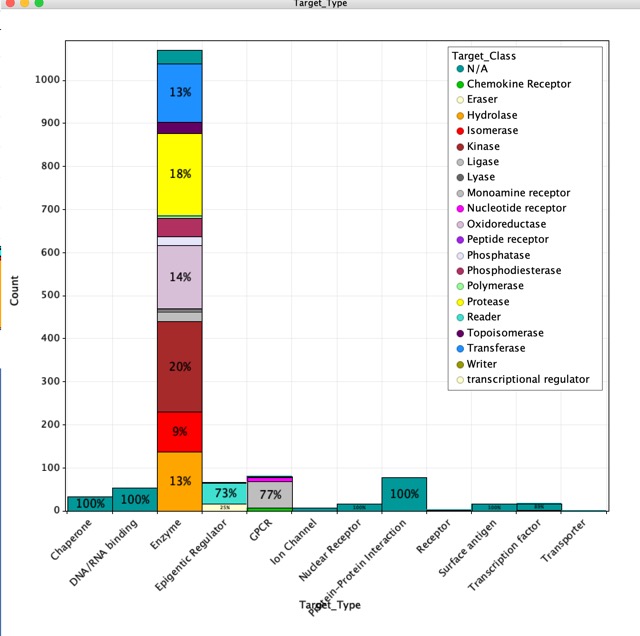
It could be argued that published fragment hits perhaps gives us an insight into the best fragments to include in library design.
BioBlocks
I've added a new entry on to the available fragments page, BioBlocks is a newcomer to the field of fragment collections. Whilst many collections are culled from available compound collections using calculated property filters (eg Rule of 3), BioBlocks have designed novel fragments and as such there is negligible overlap with other collections. One concern with bespoke fragments is that it is often a challenge to find related analogues for followup. The BioBlocks Comprehensive Fragment Library (CFL) is a subset generated from a >1 million member synthesizable virtual library, so follow up compounds can be generated using proven in house chemistry.
Practical Fragments Poll
The latest fragment-finding methods poll has been published on Practical Fragments.
The results underline the increase in the use of fragment based screening across the industry with 85% of the respondents now reporting that they actively use fragment screening. The technologies used to detect binding have also diversified with X-ray, NMR and SPR dominating. This mirrors my findings from published fragment hits. The choice of detection technology may be due to the additional structural information that X-Ray and NMR can offer.
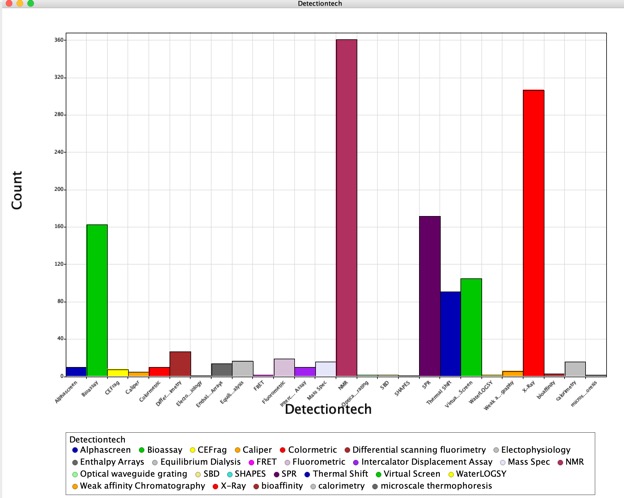
I was delighted to see this comment,
For the first time we asked about use of literature to identify fragments, and nearly a third of respondents said they incorporate previously published fragments into their work. As the amount of publicly available information continues to increase it will be interesting to see whether this number grows.
I'll be updating the published fragment hits at the end of the year.
European Lead Factory
Great news! European Lead Factory has restarted.
Pivot Park Screening Centre has successfully completed the first ultra-High Throughput Screening in IMI’s ESCulab project, and as such restarting the operations of the European Lead Factory. The screening on the European Compound Collection of ~500.000 compounds using a biochemical 1536-wells assay was finished within 4 days. Currently triaging of the UK owned program is ongoing within the Consortium, applying further biochemical and biophysical follow-up assays as well as the resynthesis of promising hits.
The programme is currently accepting proposals http://www.europeanleadfactory.eu/drug-target-assays.
European Lead Factory looking for novel screening programmes again
The European Lead Factory has been funded for another round of screening activities but under a new name European Screening Centre: unique library for attractive biology ESCulab. Over the next five years, the European Lead Factory will initiate 185 new drug discovery projects by screening medically relevant drug targets from European researchers, small and medium-sized enterprises and pharmaceutical industry against the ELF library of 550,000 unique chemical compounds.
The European Lead Factory was launched in 2013 and set up a joint collection of half a million compounds and a state-of-the-art high throughput screening centre. By the time the project ended last year, they had delivered results to researchers in universities, small biotechs and large companies across Europe, helping them to identify potential new drug candidates and breathing new life into a range of disease areas. In many cases, the seeds sown by the European Lead Factory resulted in new patents, partnering deals, and two start-ups. Now, a new IMI project, ESCulab will build on the work of the European Lead Factory. This means that researchers with drug targets can apply to screen the project’s compound collection for hits and get help developing any compounds further if they like. Jon de Vlieger, coordinator of the ESCulab consortium at Lygature, said: ‘It’s truly exciting to continue the onboarding of new and innovative proposals for screening and provide high quality starting points for drug discovery to academics and SMEs throughout Europe. In an effort to broaden our scope we are not only looking for target-based approaches, but now also enable phenotypic screens.’
You can apply here. If you don't have an assay in a format suitable for ultra high-throughput screening it is worth noting that the Wellcome Trust have small awards designed to help with the technology change required for HTS.
European Lead Factory update
The European Lead Factory (ELF) secured a total project budget of EUR 36.5 million under the second framework of the Innovative Medicines Initiative (IMI). 20 partners in 7 countries will push forward the transformation of potential drug targets to new medicines in the new project ESCulab (European Screening Centre: unique library for attractive biology) under the European Lead Factory brand.
Full details here https://www.europeanleadfactory.eu/news-events/european-lead-factory-europe’s-largest-collaborative-drug-discovery-platform-continues.
Over the next five years, the European Lead Factory will initiate 185 new drug discovery projects by screening medically relevant drug targets from European researchers, small and medium-sized enterprises and pharmaceutical industry against the ELF library of 550,000 unique chemical compounds.
Atomwise AIMS awards

I suspect many will have noticed the recent announcement of the Early Results in Drug Discovery Partnership with AI Biotech Company. These are the first results of the Atomwise AIMS awards:
The researchers have been using Atomwise’s AI-powered in silico screening technology to develop therapeutic treatments for, among others, certain types of strokes, hand-foot-and-mouth disease, and an infection that causes reproductive failure in pigs.
The AIMS award program is a great opportunity for university research scientists to easily access AI-assisted structure-based virtual screening technology:
- Customized small molecule virtual screen using AtomNet™ technology
- 72 small molecules predicted to bind to a specific target protein – QC verified by mass spectrophotometry, resuspended and diluted to a convenient concentration, aliquoted into microtiter plates, and delivered at no cost to the researcher
- Support from Atomwise’s medicinal chemists and structural biologists
- Opportunity to receive up to $30K USD to subsidize assay work
If you have a target protein with an X-ray crystal, Cryo-EM, or NMR structure, or with close sequence homology to a protein with available structures, and an assay in place to evaluate 72 potential hits, then you should consider applying.
Full details are on the AIMs awards page and the closing date is 29 April 2019.
Developing an assay for high-throughput screening
SULSA’s Assay Development Fund is actively recruiting innovative molecular targets for which there is a strong rationale for therapeutic potential.
Development of a high-throughput assay provide access to the various drug discovery initiatives that are available to the academic community, i.e. MRC DPFS, Bayer G4Targets, Wellcome Trust translational fund, the European Lead Factory, AZ innovation portal etc.
Drug Discovery Resources Updated
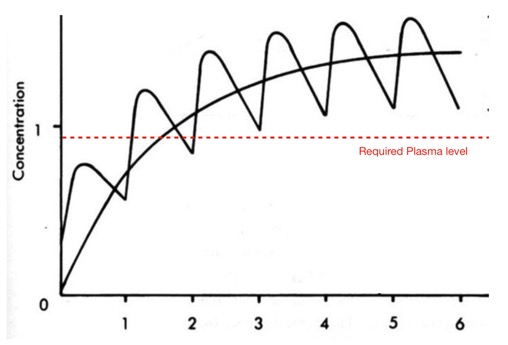
I've made a few additions and updated to the Drug Discovery Resources pages. In particular I've updated the covalent inhibitors page and added additional examples to the molecular interactions page. I've also started updating the ADME section and added a page on half-life and how it might be modulated.
Privileged Structures
The term "privileged structures" was first coined by Ben Evans DOI who recognised the potential of certain regularly occurring structural motifs as templates for derivatization to discovery novel ligands for binding to proteins. Such motifs are of course distinct from false positives molecules that appear in multiple screens due to assay interferences.
A recent publication describes a related approach looking for multi target ligands, a systematic analysis of currently available X-ray structures for compounds forming complexes with different targets DOI, by using X-ray structures they aim to avoid molecules that might interfere with the assay, some of these ligands were described in the medicinal chemistry literature, making it possible to consider additional target annotations and search for analogues. This work identified 133 unique analogue-series-based scaffolds were isolated that can serve as templates for the design of new compounds with multitarget activity.
I've added this to the Privileged Structures page of the Drug Discovery Resources.
D3R Grand Challenge 2: blind prediction of protein–ligand poses, affinity rankings, and relative binding free energies
The Drug Design Data Resource (D3R) is an NIH-funded resource dedicated to improving method development in ligand docking and scoring through community-wide blinded prediction challenges (http://www.drugdesigndata.org). DOI
The Drug Design Data Resource (D3R) ran Grand Challenge 2 (GC2) from September 2016 through February 2017. This challenge was based on a dataset of structures and affinities for the nuclear receptor farnesoid X receptor (FXR), contributed by F. Hoffmann-La Roche. The dataset contained 102 IC50 values, spanning six orders of magnitude, and 36 high-resolution co-crystal structures with representatives of four major ligand classes. Strong global participation was evident, with 49 participants submitting 262 prediction submission packages in total. Procedurally, GC2 mimicked Grand Challenge 2015 (GC2015), with a Stage 1 sub-challenge testing ligand pose prediction methods and ranking and scoring methods, and a Stage 2 sub-challenge testing only ligand ranking and scoring methods after the release of all blinded co-crystal structures. Two smaller curated sets of 18 and 15 ligands were developed to test alchemical free energy methods. This overview summarises all aspects of GC2, including the dataset details, challenge procedures, and participant results. We also consider implications for progress in the field, while highlighting methodological areas that merit continued development. Similar to GC2015, the outcome of GC2 underscores the pressing need for methods development in pose prediction, particularly for ligand scaffolds not currently represented in the Protein Data Bank (http://www.pdb.org), and in affinity ranking and scoring of bound ligands.
Conclusions:
- Successful prediction of ligand–protein poses depends on the entire workflow, including factors extrinsic to the core docking algorithm, such as the conformation of the protein selected.
- The accuracy of pose predictions tends to be improved by the use of available structural data, via ligand overlays and/or selection of receptor structures solved with similar ligands.
- The accuracy of the poses used in structure-based affinity rankings does not clearly correlate with ranking accuracy.
- Explicit solvent free energy methods did not, overall, pro-vide greater accuracy than faster, less detailed scoring methods
Selecting hits from virtual screening
Whilst high-throughput screening (HTS) has been the starting point for many successful drug discovery programs the cost of screening, the accessibility of a large diverse sample collection, or throughput of the primary assay may preclude HTS as a starting point and identification of a smaller selection of compounds with a higher probability of being a hit may be desired. Directed or Virtual screening is a computational technique used in drug discovery research designed to identify potential hits for evaluation in primary assays. It involves the rapid in silico assessment of large libraries of chemical structures in order to identify those structures that most likely to be active against a drug target. The key question is then how many molecules do you select from your virtual screen?
The results of a virtual screening run are effectively a rank ordering of the virtual screening deck ordered by whatever scoring function(s) that have been used. The task then becomes selection of molecules for experimental determination of activity.
I posed this question on the website and the results are shown below. Whilst this obviously a limited snapshot it is interesting that there is a wide variety of responses.
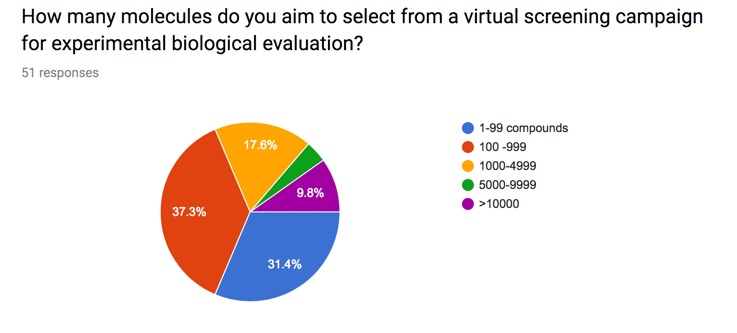
I've included the results on the page on Selecting Compounds from a Virtual Screening Run.
Trust but verify
There is an article in Nature describing a collection of problems that have arisen from incorrect chemical structures in biological screens DOI. I'm slightly surprised that this is regarded as newsworthy, but I guess it serves as a timely reminder.
The data from screening campaigns invariably contains errors
- It is often a single point assay
- Quality and diversity of Sample Collection is variable
- Compounds may interfere with the detection system
- False positives due to aggregation
- High density plates can result in cross contamination, edge effects
There is a very simple mantra you should adopt when analysing screening data "Trust but verify".
- Check compounds that were found active against the selected target are re-tested using the same assay conditions used during the HTS.
- Does a resynthesised (not repurchased) show the same activity
- Dose response curve generation: an IC50 or EC50 value is then generated, does it have a reasonable slope? Uneffected by incubation time.
- Are related analogues available, check for genuine Structure-Activity Relationships

There is a strategy for the analysis of HTS data in the Drug Discovery Resources.
MRC opens drug discovery funding applications
The Medical Research Council (MRC) recently announced that it will provide funding for up to ten high throughput screening (HTS) projects a year to run within the AstraZeneca UK Centre for Lead Discovery. The centre is home to NiCoLa-B, the world’s the world's most advanced drug discovery robot. NiCoLa-B can test up to 300,000 compounds a day
Closing date is 27 Sept 2017
https://www.mrc.ac.uk/funding/browse/mrc-az-cld/mrc-astrazeneca-centre-for-lead-discovery/.
Further information on the collaboration with AstraZeneca
Virtual Screening Pages Updated
I've updated the pages describing virtual screening, in particular the docking section.
European Lead factory "Hit to Lead" workshop
The European Lead Factory hold annual meetings intended to support early career scientists, this years meeting will focus on "Hit to Lead optimisation". The meeting will be held at Janssen Pharmaceutica NV, Beerse, Belgium, November 6th - 7th 2017.
Full details are here https://www.europeanleadfactory.eu/early-career-researcher-event/.
Free participation (incl. reimbursement of travel costs up to € 250, accommodation, access to the social get-together) at the conference.
The draft agenda is here.
A great opportunity for those just starting on their drug discovery career.
Fragment Screening - XChem at Diamond
Fragment-based screening has become increasingly popular over the last 10 years and has proven to be a viable alternative to high-throughput screening. The appeal has been driven by several features
- “Fragment Space” is smaller than “Chemical Space” and can be more effectively probed with a relatively small library
- A million compounds cover only a small fraction of the suggested 1060 Chemical Space, whilst 2000 compounds can probe much of the 106 Fragment Space
- Protein requirements should be smaller
- Binding Efficiency for small molecules is likely to be higher
- Hit rates for Fragment-based screening appear to be higher, typically 3-10%.
Whilst there are a number of biophysical methods used for Fragment-based screening structural information is often a critical step in moving the programme forward. X-ray crystallography is a very powerful technology for use in converting a "hit" into a lead for drug discovery. However, the experimental overheads have historically been too high for it to be widely used for primary screening. The X-Chem project at Diamond aims to make the technology more widely accessible.
At Diamond beamline I04-1, the full X-ray screening experiment has now been implemented as a highly streamlined process, allowing up to 1000 compounds to be screened individually in less than a week (including 36 hours' unattended beamtime). The process covers soaking, harvesting, automatic data collection, and data analysis; fragment libraries are available, though users can bring their own.
An overview of the process is available here in practice, users must generate the crystals in their home lab, and are required to come and perform soaking and harvesting themselves. Users do not need to be present for the X-ray data collection when data is collected automatically. Data analysis builds on the existing automatic data processing, and they have developed tools to streamline density interpretation and refinement (PanDDA and XChemExplorer). Use of these tools at Diamond is optional but highly recommended.
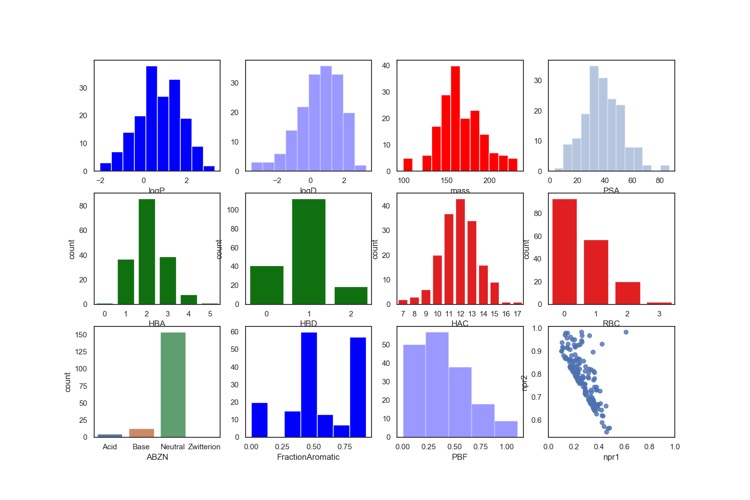
Whilst users are free to bring their own libraries a number of libraries are also available these include the Maybridge 1000, Edelris fragments, and Diamond-SGC Poised Library (DSPL), a fragment library designed to allow rapid, cheap follow-up synthesis to provide quick SAR data. The calculated physicochemical properties of the DSPL fragment collection are shown below.
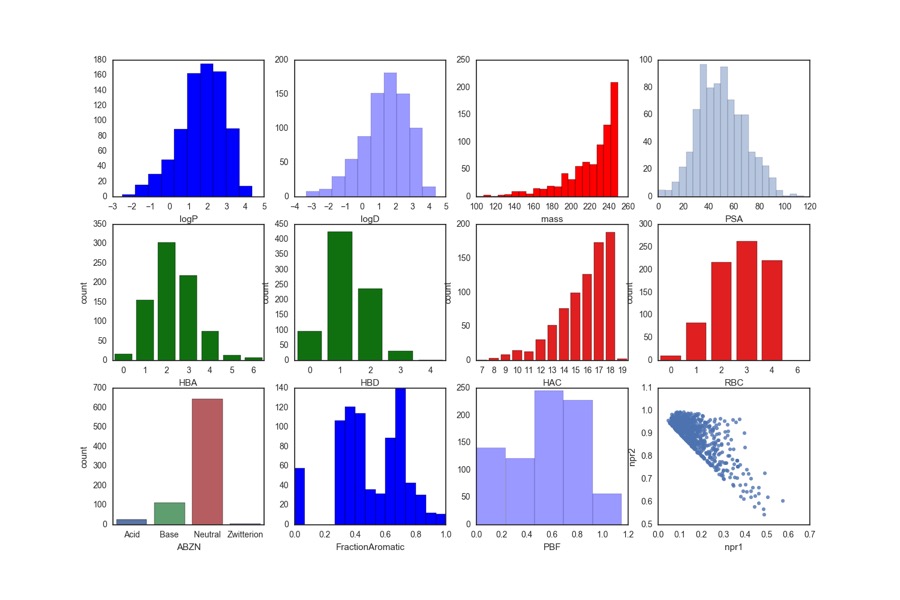
For more details on the design of the library O. B. Cox, K. Krojer, P. Collins, O. Monteiro, R. Talon, A. Bradley, O. Fedorov, J. Amin, B. D. Marsden, J. Spencer, F. von Delft, P. E. Brennan, A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chem. Sci.,2016. 7: p. 2322-2330 DOI.
There is more information in this podcast https://www.ndm.ox.ac.uk/frank-von-delft-x-rays-for-drug-discovery
The European Lead Factory Works!
The European Lead Factory (ELF) is a collaborative public-private partnership aiming to deliver novel lead molecules for drug discovery programs. When the consortium was formed around 5 years ago there was a lot of scepticism about whether a group of 30 partners rating from large Pharma companies to small academic groups could ever agree on a legal framework that would allow the ELF to function. In a addition, in an industry where confidentiality was critical to maintaining intellectual property the idea that a group of large Pharma companies would share their sample collections often regarded as the "Crown Jewels" seemed impossible. However I was at the European Lead factory Stakeholder Meeting (24-25 April 2017) and it is clear that is has been a success.
Drug Discovery Resources Updates
I've been updating the Drug Discovery Resources section.
In particular I've added a section on target prediction tools, updated the section on Analysis of HTS data, and expanded the section on aggregation in bioassays to include a recent publication giving an example of target specific aggregation.
The crystal structure showing the interaction is available (PDB 5MU8) and is displayed below using 3Dmol.js, the ligand (JNJ525) is shown in red..
Mouse Controls
| Movement | Mouse Input | Touch Input | ||
|---|---|---|---|---|
| Rotation | Primary Mouse Button | Single touch | ||
| Translation | Middle Mouse Button or Ctrl+Primary | Triple touch | ||
| Zoom | Scroll Wheel or Second Mouse Button or Shift+Primary | Pinch (double touch) | ||
| Slab | Ctrl+Second | Not Available |
In addition I've updated the section on molecular interactions.
European Lead factory Newsletter
The latest issue of the European Lead Factory Newsletter has just been published.
Highlights include the European Lead Factory recently reached the milestone of having received over 1,000 chemical library proposals for consideration. To date, 143,529 novel compounds, out of the 200,000 prospected compounds for the Public Compound Collection, have been synthesized, which means that the ELF compound collection grows daily by approximately 250 novel compounds to eventually constitute the 500,000 Joint European Compound Library (JECL).
Parkinson’s UK and the University of Sheffield have launched a joint venture biotech company, Keapstone Therapeutics. Parkinson’s UK has allocated 1 million GBP over the next sixteen months to further develop compounds that boost the internal cellular defence mechanisms against oxidative stress. These compounds were discovered by European Lead Factory.
The Ecstasy and Agony of Assay Interference Compounds
There is an editorial in ACS Central Science DOI that I would encourage everyone involved in hit identification to read.
A couple of quotes will give you an idea of the content
Alarmingly, up to 80–100% of initial hits from screening can be artefacts if appropriate control experiments are not employed.
it is important to realize that no PAINS-containing drug has ever been developed starting from a protein-reactive PAINS target-based screening hit
They also emphasise the critical need for experimental validation for any screening hit.
Such validation experiments include classic dose response curves, lack of incubation effects, imperviousness to mild reductants, and specificity versus counter-screening targets. If a molecule is flagged as a potential PAINS or aggregator using published patterns but is well-behaved by these criteria, it may be a true, well-behaved ligand. Ultimately, genuine SAR combined with careful mechanistic study provides the most convincing evidence for a specific interaction. Covalent and spectroscopic interference molecules act via specific physical mechanisms, for which controls are known. Colloidal aggregation, fortunately, is readily identified by rapid mechanistic tests and by counter-screening.
In addition you need to consider compound identify and purity, reproducing the activity with an authentic sample is essential.
Whilst time-consuming this validation work will save a fortune in the future.
Peptidyl-Prolyl cis-trans Isomerase (PPIase) assays
Having worked wit Selcia on a number of projects I always keep an eye out for news on their work on Peptidyl-Prolyl cis-trans Isomerases (PPIase). These are very interesting class of enzymes whose principal function is to catalyse the cis-trans isomerisation of the X-Pro peptide bonds in polypeptide chains (where X is any amino acid). This transformation is thought to be a mechanism to modulate protein function.
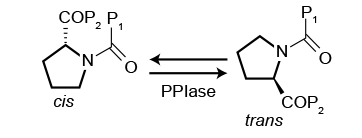
PPIase enzyme targets are of increasing interest in drug discovery due to the extensive potential of small molecule inhibitors in a range of therapeutic areas, including infection, inflammation, cancer and neuroprotection.
Selcia have now expanded the range of Peptidyl-Prolyl cis-trans Isomerase (PPIase) assays they can offer.
The SULSA Assay Development Fund:
I've worked with a couple of academic groups who have a very interesting target but no small molecule leads. Whilst there are several places offering high-throughput screening the stumbling block is often development of a robust screening assay running in a high density format. Most academic groups have little experience in developing such an assay and to be honest will need to do so very infrequently.
The SULSA Assay Development Fund: accelerating translation of new biology from academia to pharma DOI describes efforts to support the development of such assays, work carried out in collaboration with the European Lead Factory in Newhouse.
However, many scientifically interesting, novel molecular targets lack associated high-quality, robust assays suitable for hit finding and development. To bridge this gap, the Scottish Universities Life Sciences Alliance (SULSA) established a fund to develop assays to meet quality criteria such as those of the European Lead Factory. A diverse project portfolio was quickly assembled, and a review of the learnings and successful outcomes showed this fund as a new highly cost-effective model for leveraging significant follow-on resources, training early-career scientists and establishing a culture of translational drug discovery in the academic community.
European Lead Factory technical acceptance criteria
- Minimally 384 wells (max 30 μl)
- Homogenous assay (no washing steps)
- Defined endpoint
- Z-prime >0.6
- Readout stability >1 h
- S/B signal >3
- Incubation times <4 h
- DMSO tolerance: minimum 0.5%
- All reagents minimally 8 h stable
- Recombinant proteins(s) >80% pure
Perhaps this quote sums things up very nicely.
‘We only got so far in optimising our assay because in my opinion we do not have either the equipment or the personnel with the skill set required to do this efficiently. So being able to come and work at Newhouse where you are then surrounded by people who can identify quickly where assay improvements should be made and then pass on the knowledge of how to set about applying these changes (as well as gaining experience with various pieces of equipment) was invaluable.’
More details are available here http://www.sulsa.ac.uk/sites/sbsweb2.bio.ed.ac.uk.sulsa/files/downloads/SULSAassaydevelopmentfundguidancenotes4.pdf
The Wellcome Trust also provides Seed/Pathfinder awards that could cover similar projects.
Another European Lead Factory success
We are now starting to see some of the results of the screening of academic projects at the European Lead factory that was initiated in 2013.
Dr Mahlapuu’s group, based at the University of Gothenburg, first identified a new target which could be used to reverse metabolic complications in type 2 diabetes. With the help of the European Lead Factory experts, she then screened the Joint European Compound Library of the then 320,000 industry compounds and identified a set of selective and potent small molecules which interfere with this target.
They have now formed a spinout company to develop these leads. ScandiCure has received 1 MSEK from the 2016 SWElife program to continue the development of first-in-class anti-diabetic drug based on small molecule antagonists of a novel key mediator - serine/threonine protein kinase 25 (STK25).
ELF and antibiotic resistance programme
The latest news report from the European Lead Factory highlights work targeting antimicrobial resistance in collaboration with Professor Chris Schofield (University of Oxford). The high throughput screen of >300,000 compounds and initial triaging provided 50 qualified hits.
Multiple series of compounds were validated through resynthesis, biochemical and biophysical profiling at the European Screening Centre site in Newhouse, complemented with ligand–protein crystallography and antimicrobial evaluation at University of Oxford
Great to see projects like this move forward, demonstrates how important the ELF is in providing hits for academic groups/small companies.
European Lead Factory Update
The latest newsletter from the European Lead Factory has just been published and highlights a number of notable achievements.
As a result of the screening campaigns >3000 qualified hits have been awarded to private and public target owners. 72 public target programmes have been accepted, 48 high throughput screens finished and 41 hit lists with associated data reports handed over to the target owners. >150 bespoke assays have been developed in order to extract the most interesting hits for public programmes.
There are now 450,000 compounds in the compound library of which 120,000 are novel compound specifically synthesised for the ELF.
You can read more details here https://www.europeanleadfactory.eu/results/
The European Lead Factory is a collaborative public-private partnership aiming to deliver innovative drug discovery starting points. Having established the first European Compound Library and the first European Screening Centre, the EU Lead Factory aims to give free access to up to 500,000 novel compounds, a unique industry-standard uHTS platform, and much more.
We are now starting to see publications describing these endeavours..
https://www.europeanleadfactory.eu/results/publications/scientific-articles/
European Lead Factory
The European Lead Factory started work just over 3 years ago and have just released an update on results to date.
The EU Lead Factory is an IMI-funded project that aims to create new chemistry based on crowd-sourced ideas and boost applicants’ drug discovery programmes at no upfront costs
- A Sweden-based start-up company, ScandiCure AB, has been founded based on the results of an EU Lead Factory target programme. ScandiCure is developed with the support and investment of GU Ventures, which is wholly owned by the Swedish State.
- Patents on EU Lead Factory compounds for treatment of multi-resistant bacteria infections and cancer.
- An EU Lead Factory programme accepted by IMI’s ENABLE for preclinical development.
- In vivo proof-of-concept generated with EU Lead Factory compounds.
- 2 PhD thesis enhanced with EU Lead Factory target programme assay development and screening results.
- >35 scientific, peer-reviewed articles, whereof one is the 4th most downloaded article published in Drug Discovery Today in 2015.
- 450,000 compounds in the Joint European Compound Library (JECL), whereof >120,000 of the prospected 200,000 novel screening compounds have been synthesised. 330,000 compounds were selected and assembled from the EFPIA participants’ proprietary compound collections within 6 months of operation.
- Many testimonials of the high quality of the EU Lead Factory output and the JECL compounds. For example, 17/49 EFPIA partner screens have triggered further work.
- 72 public target programmes accepted, 48 high throughput screens finished and 41 hit lists with associated data reports handed over to the target owners.
- In total, 2925 qualified hits have been granted public and private target owners (1041 and 1884, respectively).
- >1500 bespoke compounds synthesised in hit validation and hit-to-lead phase of public target programmes.
- >10 crystal structures of target–compound complexes have been solved.
- >150 bespoke assays have been developed in order to extract the most interesting hits for public programmes.
- 12/14 of the public target programmes offered to the industry EFPIA participants have been asked to provide a dossier for further assessment.
- >30 academic postdoctoral fellows trained in industry methods and approaches.
- Researchers in 13 countries (BE, DE, DK, ES , FR , IT, IL, HU, NL, PL , PT , SE, UK) involved. Partners spread over 8 countries and target owners over 11.
- 2 Custom-built data management platforms, the Honest Data Broker system developed to enable screening data management and triaging, whilst ensuring confidentiality and patentability; TarosGate2, a chemistry workflow management system with a secure built-in electronic laboratory notebook.
I’ve been involved in a number of screens and we have been very happy with the results, this is a great resource long may it continue.
It is also worth noting that the next submission deadline is 10th October 2016, here is the link for submitting assays for consideration. https://www.europeanleadfactory.eu/how-to-submit/drug-target-assays/
Fragment based screening
I’ve just updated some of the fragment based screening pages, in particular I’ve updated the section on Published fragment Hits. The database now contains 1216 entries culled from over 240 publications directed at nearly 174 different molecular targets using 26 different detection technologies.
I also noticed that a fragment library I was helped design is now commercially available, The Selcia Fragment Library was designed to have broad applicability and chemical tractability. It is also one of the few libraries where solubility has been confirmed experimentally. The profile of the library is included in the fragment library profiles.
Drug Discovery Resources Updated
I have updated the Drug Discovery section on Screening Collection design.
Drug Discovery Resources Update
I've updated the hit identification section of the Drug Discovery Resources. In particular I've added to the high-throughput screening analysis including more information on PAINS (Pan Assay Interference Compounds) first described by Baell et al DOI and subsequently summarised in an excellent Nature comment.
Academic researchers, drawn into drug discovery without appropriate guidance, are doing muddled science. When biologists identify a protein that contributes to disease, they hunt for chemical compounds that bind to the protein and affect its activity. A typical assay screens many thousands of chemicals. ‘Hits’ become tools for studying the disease, as well as starting points in the hunt for treatments.
These molecules — pan-assay interference compounds, or PAINS — have defined structures, covering several classes of compound. But biologists and inexperienced chemists rarely recognize them. Instead, such compounds are reported as having promising activity against a wide variety of proteins. Time and research money are consequently wasted in attempts to optimize the activity of these compounds. Chemists make multiple analogues of apparent hits hoping to improve the ‘fit’ between protein and compound. Meanwhile, true hits with real potential are neglected.
Also added a page on Aggregators. Promiscuous inhibition caused by small molecule aggregation is a major source of false positive results in high-throughput screening. To mitigate this, use of a nonionic detergent such as Triton X-100 or Tween-80 has been studied, which can disrupt aggregates, and is now common in screening campaigns DOI.
An Aggregation Advisor for Ligand Discovery
Aggregation is a regular concern when evaluating potential hits from screening and a recent paper "An Aggregation Advisor for Ligand Discovery" DOI attempts to provide an insight into this phenomenon, in addition they provide a useful web-based tool http://advisor.bkslab.org that provides a free service to advise whether molecules may aggregate under biological assay conditions.
European Lead Factory
p>The European Lead Factory has just announced that an additional 50,000 new compounds have been added to their screening collection. This brings the collection up to 350,000 compounds and sets them well on the way to their 500,000 target.
I've been involved in a couple of projects that have made use of this high-throughput screening facility and I've been impressed with the quality and diversity of the hits generated.
The European Lead Factory was established to promote the discovery of novel lead compounds, suitable for subsequent optimization either to drug candidates or to high‐quality pharmacological tools for the experimental validation of targets.
If you have a target you want to screen against you can submit a proposal online. For an academic or small company this is an interesting way to identify novel starting points for a medicinal chemistry program.
Free Compound Screening for antimicrobial activity
The concerns about antibiotic resistance are well known and indeed have made headlines in the mainstream press. Here is a chance to help find the next generation of antibiotics.
Do you have interesting compounds sitting on the shelf? Perhaps you would be interested in having them screened for antibiotic activity for free?
The Community for Open Antimicrobial Drug Discovery would like to hear from you, their goal is to screen compounds from academic research groups from anywhere in the world for free.
The requirements are pretty minimal
We ask for 1-2 mg of pure compound which will be used for primary screening, hit confirmation, and if active will be used for a broader antimicrobial screening, cytotoxicity and a check for its purity. We require all compounds to be soluble in water or DMSO and to be shipped as dry material in appropriate containers, such as 1-2 mL Eppendorf tubes. For larger collections we can arrange plates or tube-racks.
In the primary screen they test against against key ESKAPE pathogens, E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, S. aureus (MRSA), as well as the fungi C. neoformans and C. albicans. The ‘ESKAPE’ pathogens that are responsible for two-thirds of all health care-associated infections and resistant strains of these bacteria represent the greatest unmet need in antibacterial drug development.
New Compound Sets Identified from High Throughput Phenotypic Screening Against Three Kinetoplastid Parasites: An Open Resource
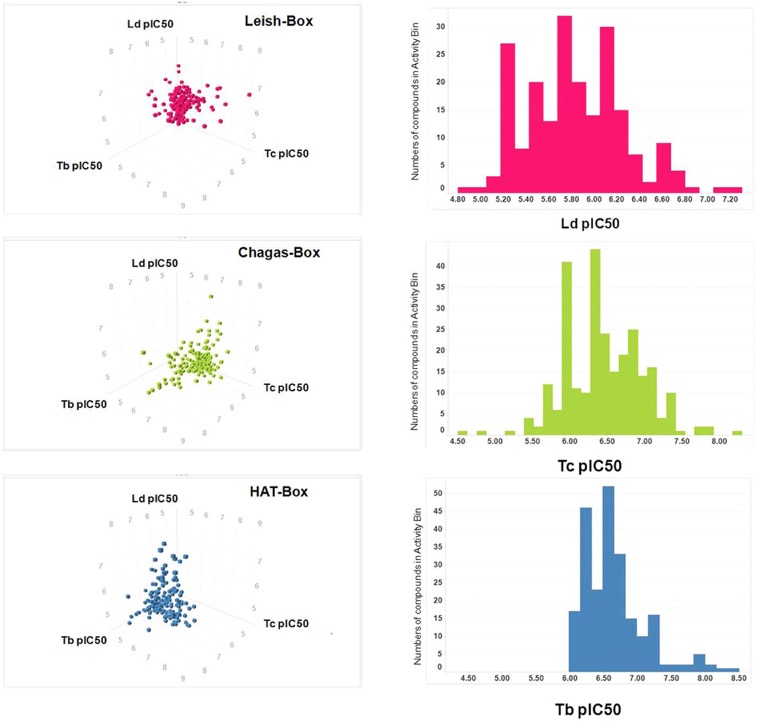
The GSK high-throughput screening group at Tres Cantos and collaborators have just published DOI the results of whole-cell phenotypic screens against the three kinetoplastids most relevant to human disease, i.e. Leishmania donovani, Trypanosoma cruzi and Trypanosoma brucei. Three anti-kinetoplastid chemical boxes of ~200 compounds each were assembled. Functional analyses of these compounds suggest a wide array of potential modes of action against kinetoplastid kinases, proteases and cytochromes as well as potential host-pathogen targets. The compound sets are provided as an open resource for the scientific community.
More on PAINS
I often get asked to help with the analysis of high-throughput screening results and one of the first filters I run as part of the hit identification is to flag for PAINS (Pan Assay Interference Compounds) first described by Baell et al DOI and subsequently summarised in an excellent Nature comment.
Academic researchers, drawn into drug discovery without appropriate guidance, are doing muddled science. When biologists identify a protein that contributes to disease, they hunt for chemical compounds that bind to the protein and affect its activity. A typical assay screens many thousands of chemicals. ‘Hits’ become tools for studying the disease, as well as starting points in the hunt for treatments.
These molecules — pan-assay interference compounds, or PAINS — have defined structures, covering several classes of compound. But biologists and inexperienced chemists rarely recognize them. Instead, such compounds are reported as having promising activity against a wide variety of proteins. Time and research money are consequently wasted in attempts to optimize the activity of these compounds. Chemists make multiple analogues of apparent hits hoping to improve the ‘fit’ between protein and compound. Meanwhile, true hits with real potential are neglected.
In the supplementary information they provided the corresponding filters in Sybyl Line Notation (SLN) format, however they have also been converted to SMARTS format and incorporated in sieve file for use in filtering compound collections. If you are a Vortex user then there is also a Vortex script available, filters are also available for Knime and now it is even available on mobile devices with MolPrime+.
It is probably not until you have been involved in multiple small molecule screens that you appreciate the number of ways that false positives can occur and just how much valuable time and resources can be wasted following them up. Indeed it may be for the more difficult targets the majority of hits seen may be false positives. Flagging PAINS is now such a well developed tool that it would be fool hardy not to include it.
Drug Discovery Resources Updates
I’ve made a couple of updates to the Drug Discovery Resources pages. In particular I’ve updated the Published fragments Hits to include more examples, details of “promiscuous” compounds and summary of detection technologies and the targets explored. I’ve also updated the Aspartic Protease inhibitors page.
As ever comments and/or suggestions very welcome.
Kinase Inhibitors
I’ve updated the Drug Discovery Resources to include a page on Kinase Inhibitors. I will be expanding it over the next week, so any comments or suggestions welcome.