Hit Finding Strategies
“The single most important factor determining the likelihood of success of a project is the quality of the starting lead”, Anon
In an analysis of 156 published clinical candidates from the Journal of Medicinal Chemistry between 2018 and 2021 "An Analysis of Successful Hit-to-Clinical Candidate Pairs?" DOI the source of the initial hit was identified.. The results are shown in the plot below
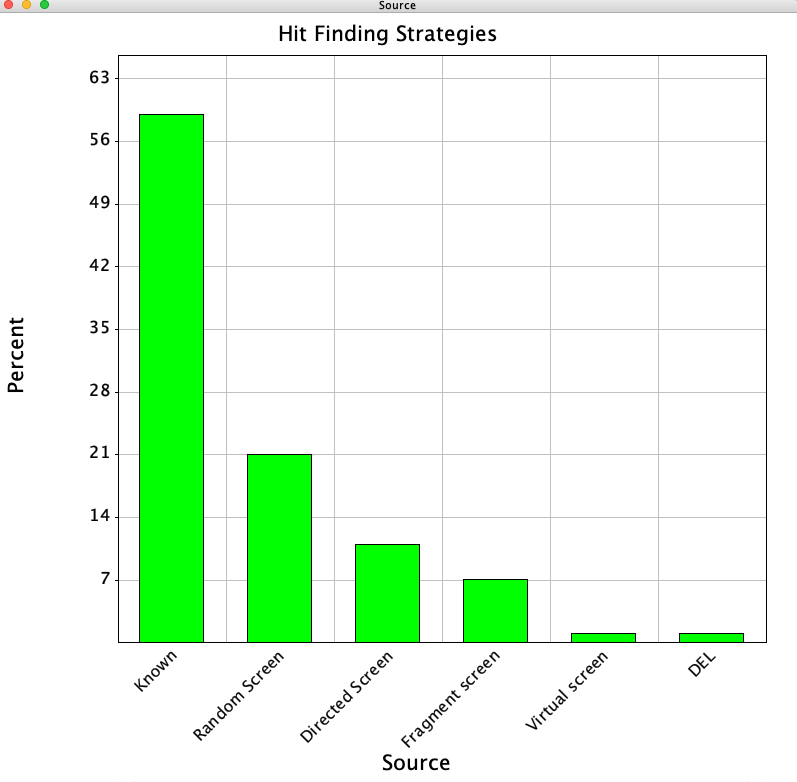
They categories the lead sources into 6 categories depending on the initial hit-finding strategy.
- Known, this might be the endogenous ligand or a molecule taken from the published literature or patents.
- Random Screen, usually a high-throughput screen of a large compound collection.
- Structure-based drug design (SBDD), in silico screening of compound collections, including the use of the target protein 3D structure.
- Directed Screen, screening of smaller sets of compounds which are selected based on prior knowledge of the target or chemical class. Also known as focused, targeted or biased screening.
- Fragment Screen, typically libraries with a few thousand compounds or less of low molecular weights (<200 Da), screened at high concentration
- DNA encoded library, screening of very large collections (108) of small molecule compounds, using a technology that involves the conjugation of molecule to a DNA tag.
The popularity of using known molecules as starting points might at first seem surprising but this will include examples where the aim is to reduce some unexpected off-target activity/toxicity or drug-drug interaction, respond to resistance due to mutations in the target protein, or combine two different biological activities into a single molecule. Interestingly despite much recent interest there appear to be few examples of a phenotypic screen used to find hits. The discovery of rusavir (MK-8408) a HCV NS5A inhibitor DOI is one example but It is worth noting this comment in the publication "The exact mechanism of NS5A inhibition remains unclear and is poorly understood".
The analysis also highlighted the distribution of target classes with Kinases (31%) being the most popular followed by other enzymes (28%) , GPCR (10%) and Ion Channels (5%). Emerging areas highlighted include protein-protein interactions and epigenetic targets both target areas include many with open shallow binding sites requiring lager molecules to achieve high affinity binding.
I calculated the physicochemical properties of both the hits and the clinical candidates.
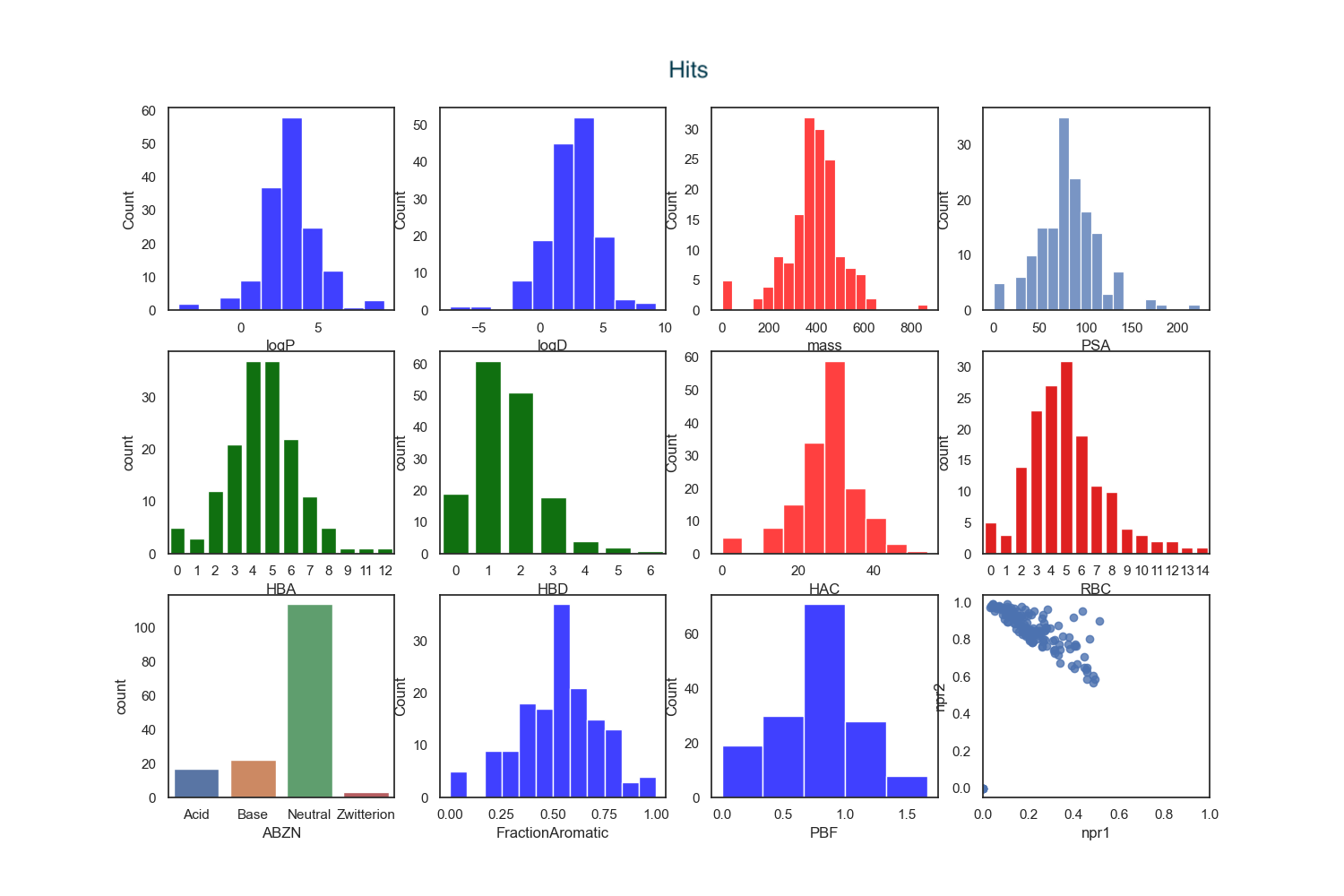
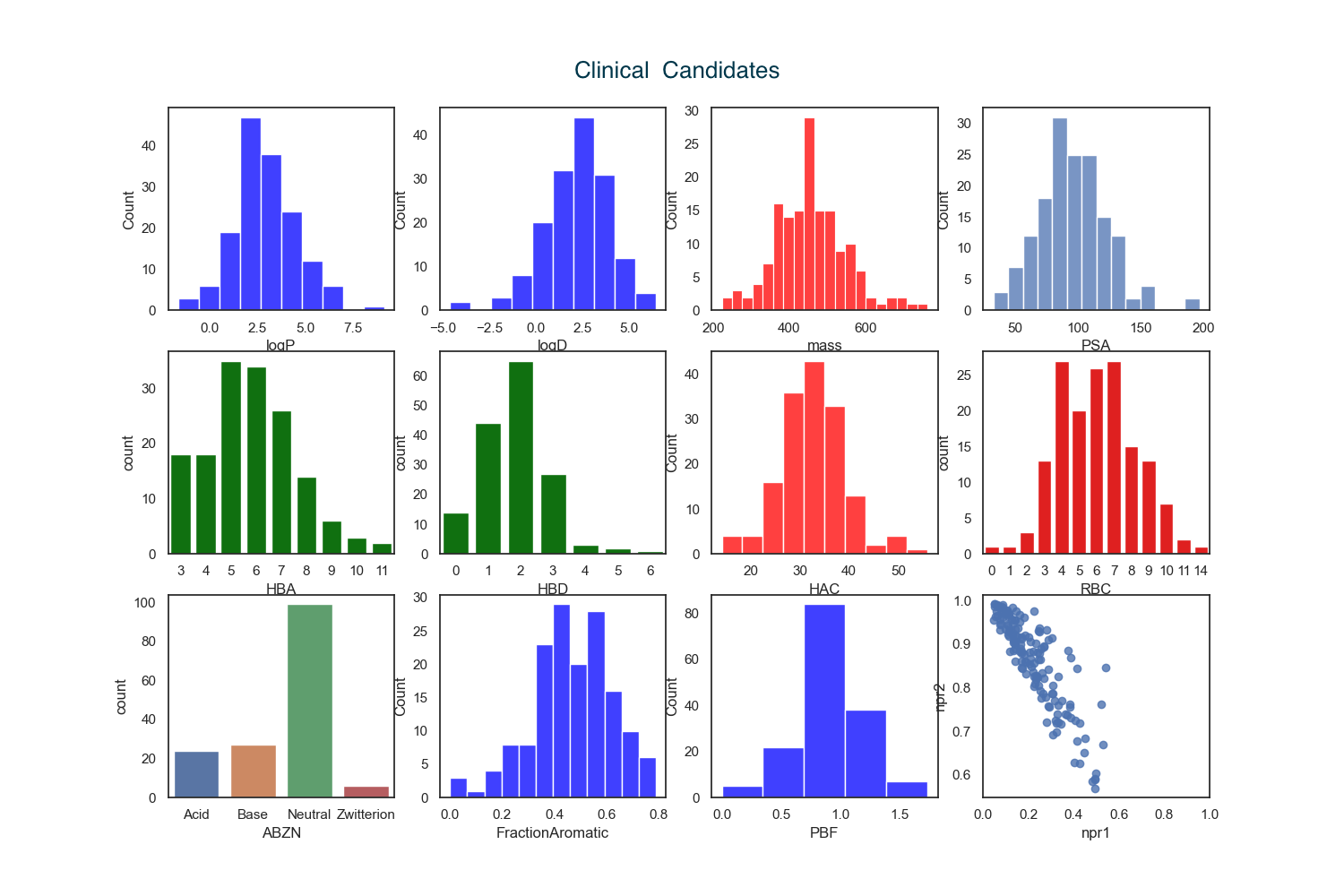
An analysis of physicochemical properties on the hit-to-clinical pairs shows an average increase in molecular weight (ΔMW = +85) but little change in lipophilicity (ΔclogP = −0.3), although exceptions are noted. Interestingly the number containing ionisable groups has increased as has the number of HBD. The majority (>50%) of clinical candidates were found to be structurally very different from their starting point and were more complex.
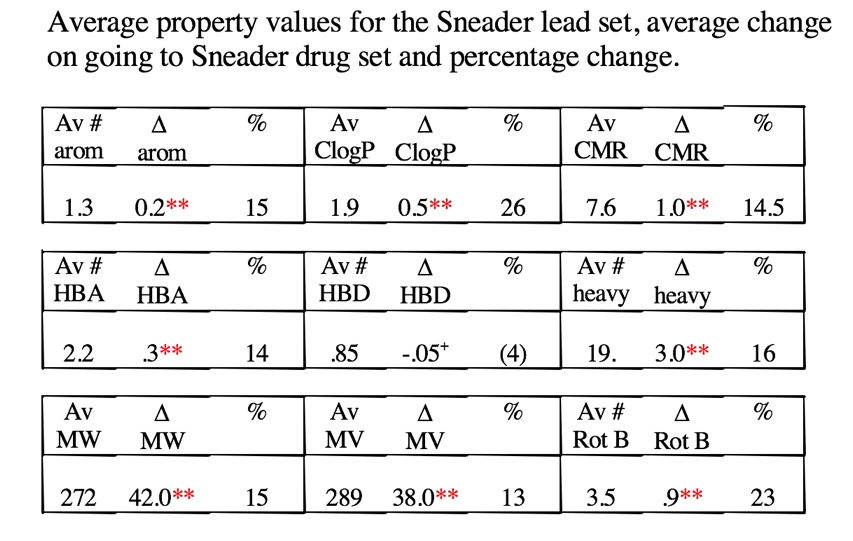
This comparison of hit to drug pairs largely mirrors the results from an analysis of W. Sneader’s book “Drug Prototypes and their exploitation" DOI with data from 480 case histories shown below.
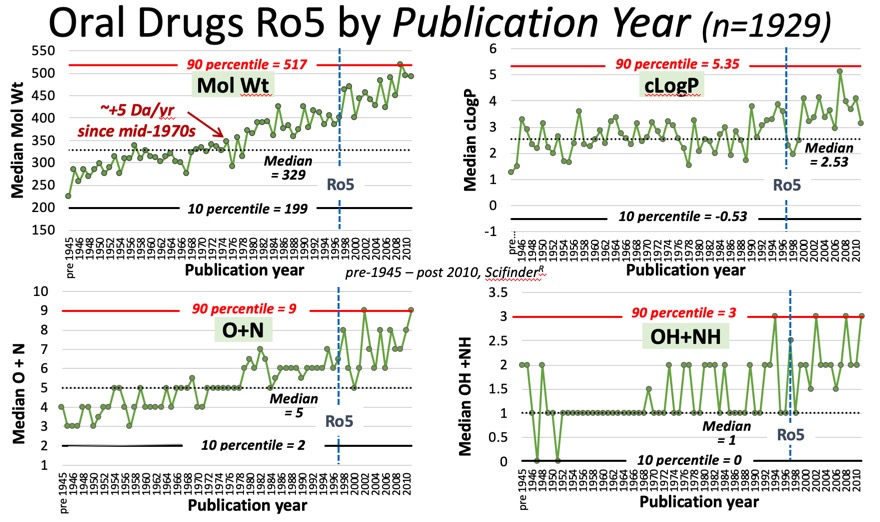
These trends appear to be continuing, looking at the properties of published drugs (we of course don’t know about all failures) since the publication of the rule of 5 paper, Molecular weight has continued to increase, cLogP appears to have plateaued around 4, whilst there is an increase in the number of HBA, there is only a marginal increase in HBD
Hit identification
The Hit confirmation phase is follows:
- Exclusion of hits with potential reactivity, assay interference or aggregation
- Re-testing: compounds that were found active against the selected target are re-tested using the same assay conditions used during the HTS.
- Dose response curve generation: an IC50 or EC50 value is then generated
- Are related analogues available, check for genuine Structure-Activity Relationships
- Check for irreversible binding
- Orthogonal testing: Confirmed hits are assayed using a different assay which is usually closer to the target physiological condition or using a different technology.
- Secondary screening: Confirmed hits are tested in a functional assay (agonist/antagonist) or in a cellular environment.
- Assessment of drug-like properties using computational analysis and early physicochemical and ADME measurements
- Chemical tractability: Medicinal chemists will evaluate compounds according to their synthesis feasibility and flexibilty towards chemical diversification or library synthesis.
- Intellectual Property evaluation: Hit compound structures are quickly checked in specialized databases to define patentability and freedom to operate.
- Hit ranking and clustering, preliminary SAR.
Building up a sample collection for High-throughput screening is a major undertaking and for a small company or academic group submitting a proposal to the European Lead Factory might be an attractive alternative. I've written a review of the ELF here.
There is an editorial in ACS Central Science DOI that I would encourage everyone involved in hit identification to read.
A couple of quotes will give you an idea of the content
Alarmingly, up to 80–100% of initial hits from screening can be artefacts if appropriate control experiments are not employed.
it is important to realise that no PAINS-containing drug has ever been developed starting from a protein-reactive PAINS target-based screening hit
They also emphasise the critical need for experimental validation for any screening hit.
Such validation experiments include classic dose response curves, lack of incubation effects, imperviousness to mild reductants, and specificity versus counter-screening targets. If a molecule is flagged as a potential PAINS or aggregator using published patterns but is well-behaved by these criteria, it may be a true, well-behaved ligand. Ultimately, genuine SAR combined with careful mechanistic study provides the most convincing evidence for a specific interaction. Covalent and spectroscopic interference molecules act via specific physical mechanisms, for which controls are known. Colloidal aggregation, fortunately, is readily identified by rapid mechanistic tests and by counter-screening.
In addition you need to consider compound identify and purity, reproducing the activity with an authentic sample is essential.
Whilst time-consuming this validation work will save a fortune in the future.
Updated 25 May 2023