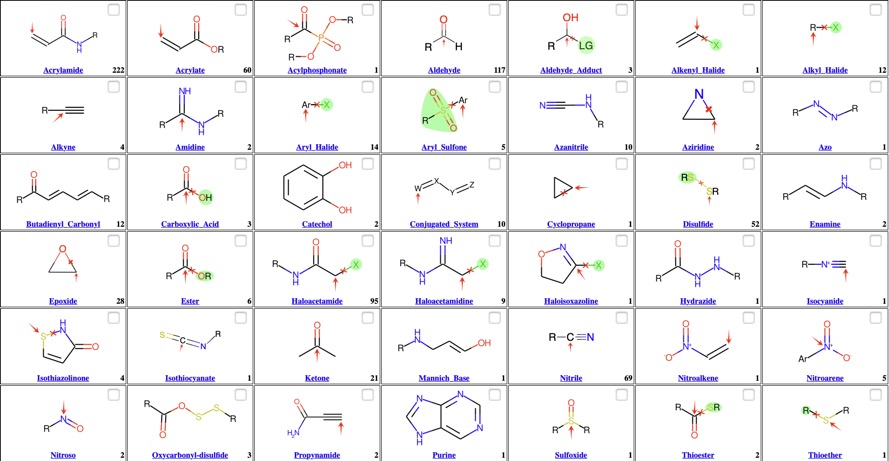
Cysteine Protease Inhibitors
Cysteine proteases have a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic dyad. The first step is deprotonation of a thiol in the enzyme's active site by a histidine residue. The next step is nucleophilic attack by the deprotonated cysteine's anionic sulfur on the peptide carbonyl carbon. In this step, a fragment of the substrate is released with an amine terminus, the histidine residue in the protease is restored to its deprotonated form, and a thioester intermediate linking the new carboxy-terminus of the substrate to the cysteine thiol is formed. The thioester bond is subsequently hydrolyzed to generate a carboxylic acid moiety on the remaining substrate fragment, while regenerating the free enzyme.
For more information on the mechanism have a look at the MACiE database (Mechanism, Annotation and Classification in Enzymes), papain PDB entry 1PAD is an example of an cysteine protease.
Inhibitors
As might be expected an enzyme with a nucleophilic sulphur in the active site a variety of electrophiles have been shown to be potent inhibitors. CovBinderDB DOI is a fantastic resource mined from the PDB that contains 7375 covalent modifications in which 2189 unique covalent binders target nine types of amino acid residues (Cys, Lys, Ser, Asp, Glu, His, Met, Thr, and Tyr) from 3555 complex structures of 1170 unique protein chains. The database can be accessed here https://yzhang.hpc.nyu.edu/CovBinderInPDB/. As you might expect Cysteine is the most common covalently bound amino acid. The different warheads used to react with cysteine are shown below.
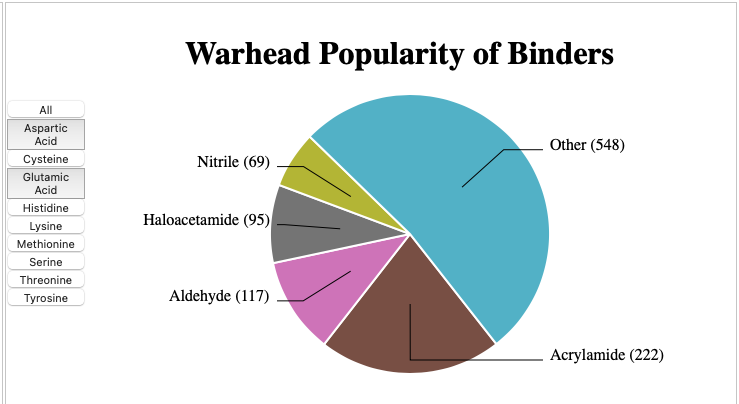
Some of the more commonly used warheads are shown below.
There are many other examples of covalent inhibitors described here
Updated 18 Jan 2023