Selecting hits from virtual screening
Whilst high-throughput screening (HTS) has been the starting point for many successful drug discovery programs the cost of screening, the accessibility of a large diverse sample collection, or throughput of the primary assay may preclude HTS as a starting point and identification of a smaller selection of compounds with a higher probability of being a hit may be desired. Directed or Virtual screening is a computational technique used in drug discovery research designed to identify potential hits for evaluation in primary assays. It involves the rapid in silico assessment of large libraries of chemical structures in order to identify those structures that most likely to be active against a drug target. The key question is then how many molecules do you select from your virtual screen?
The results of a virtual screening run are effectively a rank ordering of the virtual screening deck ordered by whatever scoring function(s) that have been used. The task then becomes selection of molecules for experimental determination of activity.
I posed this question on the website and the results are shown below. Whilst this obviously a limited snapshot it is interesting that there is a wide variety of responses.
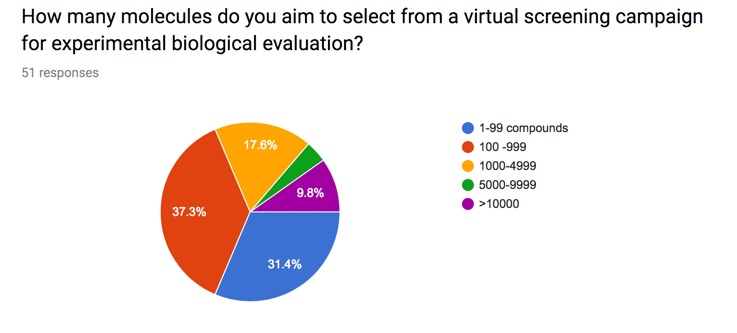
I've included the results on the page on Selecting Compounds from a Virtual Screening Run.