Profiles of Fragment Collections
A number of companies were kind enough to provide me with a file containing the chemical structures of their fragment collections for comparison. The libraries arrived in different file formats and were all converted to sdf and mdb formats using iBabel and MOE and the “wash” command in MOE used to clean up the collections and standardise protonation etc. this resulted in a total of over 160,000 available fragments.
The first step was to compare each of the libraries with each other to examine the degree of overlap between the libraries. A SVL (Scientific Vector Language) script was used: dbnbmols_incommon.svl (available from Chemical Computing Group) which calculated a matrix for the number of compounds each database has in common (or similar to) with each of the other databases based on either structure (SMILES) or fingerprints.
To look for identical structures the all the databases were opened in MOE and the SVL script loaded. Using the following SVL command,
db_nb_mols_incommon [dbv_KeyList[], 'MatrixOut.txt', [write_cpds:1, cpds_outfile:'ident_cpds.txt']]
Generated the matrix shown in Table 1 and a text file giving a list of all compounds (in SMILES format) that appeared in multiple databases and a list of databases they were present in. First looking for identical compounds in each database it is clear there is relatively little overlap. The results are shown in Table 1. The numbers in red are the number of unique compounds in each library.
Updated: A couple of companies have sent me updated files (many thanks) and I have redone the analysis
Click on the image below to open the table in a larger window.
Given the number of compounds involved it is perhaps surprising there are less than 200 compounds that are present in four or more collections some of which are shown below.

If we next use the script to look at compound similarity using Maccs fingerprints and a Tanimoto coefficient of 0.85, the results are shown in Table 2, using the SVL command,
db_nb_mols_incommon [dbv_KeyList[], 'sim_85_MatrixOut.txt', [fp_code:'FP:MACCS', sim_code:'tanimoto', thresh:0.85, fp_flag:1]]
Here you can see the different fragment collections do appear to contain some similar compounds, looking at the structures of some of the compounds it appears they usually contain privileged structures with slightly different decoration. There are a couple of things to consider when building a fragment collection. Whilst there may be some attraction to selecting fragments that are unrelated to other compounds, the downside is that it is more difficult to follow up a hit be selecting readily available commercial fragments. The “SAR by catalogue” approach can certainly give initial support to a program without the need to commit significant chemistry resources.
Click on the table below to open in a new window,
Click here to browse the physicochemical profile of each the fragment collections. A profile of the physicochemical properties (HBD, HBA, PSA, HAC, LogP, LogD, MWt, RBC) was generated using an Applescript that uses evaluate from ChemAxon to calculate the physicochemical properties and Aabel to construct the histograms. I also used it to determine pKa in order to identify acidic or basic groups and categorized the fragments accordingly, in addition I calculated the fraction of aromatic atoms (number of aromatic atoms/number of heavy atoms) since there has been concern about the number of aromatic compounds in fragment collections. I’ve also included npri (Normalized ratio of principle moments of inertia) as described by Sauer WH, Schwarz MK (2003) Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J Chem Inf Comput Sci 43:987–10030. DOI this was calculated using MOE.
My browsing suggested a number of common scaffolds so I used the sca.svl script within MOE to find all the scaffolds in the combined collections. If we ignore the simple phenyl rings then a number of scaffolds appear regularly, and are shown below.

A profile of the physicochemical properties (HBD, HBA, PSA, HAC, LogP, LogD, MWt, RBC) was generated using an Applescript that uses evaluate to calculate the physicochemical properties and Aabel to construct the histograms. I also used it to determine pKa in order to identify acidic or basic groups and categorized the fragments accordingly, in addition I calculated the fraction of aromatic atoms (number of aromatic atoms/number of heavy atoms) since there has been concern about the number of aromatic compounds in fragment collections. I’ve also added npmi (Normalized ratio of principle moments of inertia) as described by Sauer WH, Schwarz MK (2003) Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J Chem Inf Comput Sci 43:987–10030. DOI
Click here to browse the physicochemical profile of each the fragment collections.
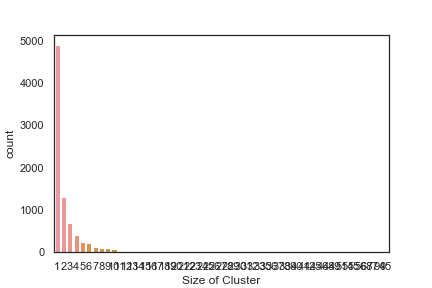
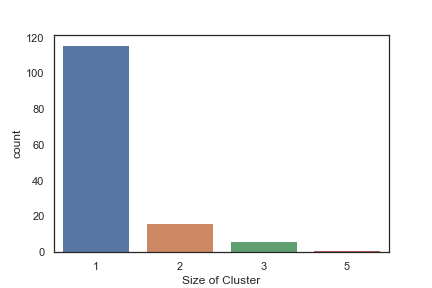
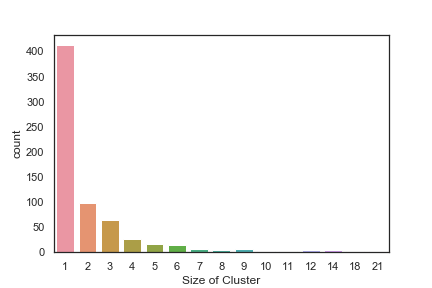
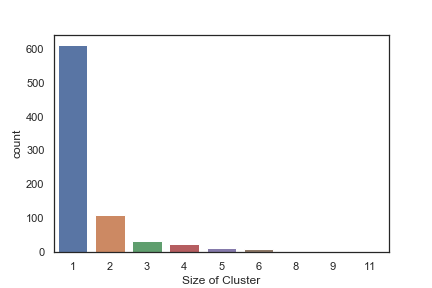
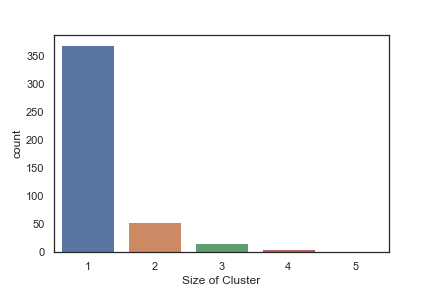
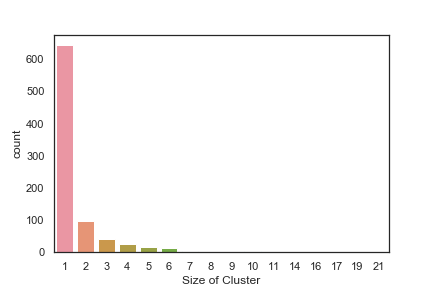
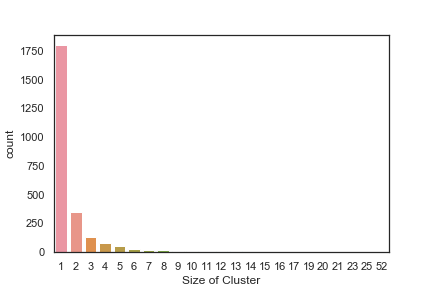
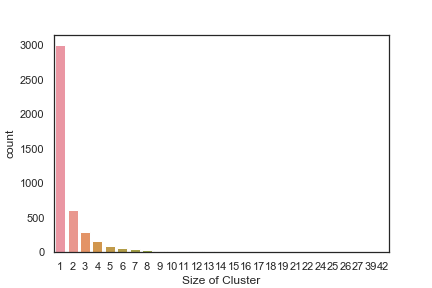
In order to get some idea of the diversity of the collections I clustered them in MOE using MACCS fingerprints and a Tanimoto cutoff of 0.85, the charts were then created in Aabel. Whilst the majority of the compounds are singletons it is clear that some of the collections do contain a few large clusters (the giveaway is the numbers on the X-axis!). I’ve also calculated the diversity coefficient for each library based on this reference D. B. Turner, S. M. Tyrrell, and P. Willett, J. Chem. Inf. Comput. Sci., 1997, 37, 18 22 using MACCS descriptors and the lib_div SVL script from Chemical Computing Group, so for Asinex the diversity coefficient is 0.63. Whilst the diversity of the screening collection is important, bear in mind that hit expansion is much easier if similar structures are readily and/or commercially available.
 | |
 |  |
 |  |
 |  |
 |  |
 | cluster.png) |
 |  |
 |  |
 | 
|
I’d be delighted to add further fragment collections.
Updated 5 December 2019

