Predicting Metabolism
All drugs are subject to metabolic process, in general these process serve to increase the polarity of molecules in an effort to increase excretion. Whilst the impact of metabolism on the drug duration of action is major concern, a knowledge of the metabolic routes can be useful in other ways. In particular, knowledge of the mechanism may highlight potential high energy intermediates that could contribute to toxicity, the identification of metabolites that in themselves may or may not have pharmacological effects, changes in physicochemical properties resulting from biotransformation.
Several approaches have been used to produce in silico systems to predict metabolism,
Local solutions, intended to predict the activity of a single enzyme (and often only within a chemical series). These models can be based on pharmacophores (QSAR) derived from known substrates, or docking potential substrates into the active site of the enzyme, and/or quantum mechanical calculations used to predict reactivity. The majority of drug are metabolised by Cytochome P450 enzymes that exist predominantly in the liver. Whilst similar in structure the CYPs have distinct substrate specificities and models for each of the enzymes need to be derived. The recent crystallization of CYP P450s should help refine these models. A number of the computational models for predicting CYP mediated metabolism have been reviewed (European Journal of Medicinal Chemistry Volume 41, Issue 7, July 2006, Pages 795-808).
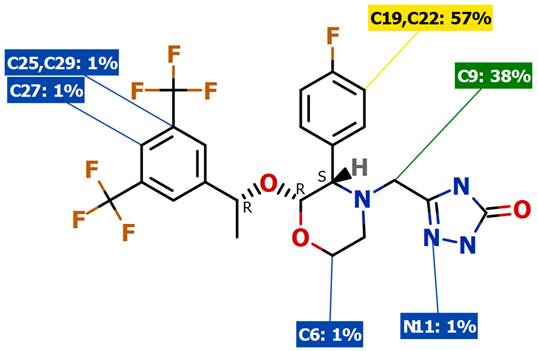
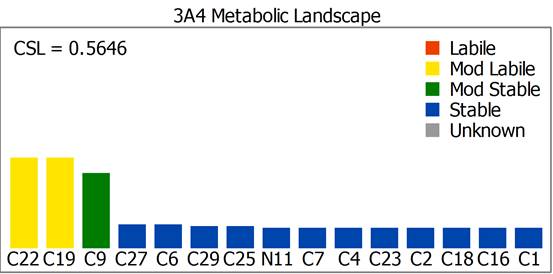
SMARTCyp is a method for prediction of which sites in a molecule that are labile for metabolism by Cytochromes P450 isoform 3A4. It is also a reactivity model which is applicable to all P450 isoforms. The method has been published as SMARTCyp – a 2D-method for Prediction of Cytochrome P450 Mediated Drug Metabolism in ACS Medicinal Chemistry Letters, DOI. The results for the sites of metabolism for the NK1 antagonist Emend are shown below.
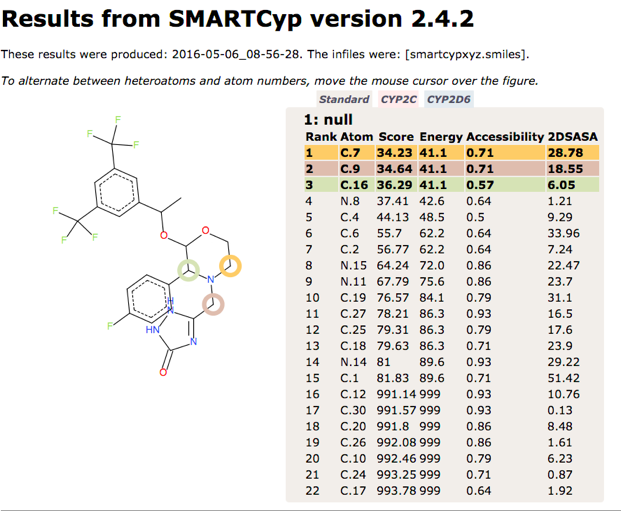
The SMARTCyp methodology has been used by several groups to enhance other tools, RS-Predictor DOI. Combination of RS-Predictor and SMARTCyp are shown to have stronger performance than either method alone. RS-Predictor provides a predictive models for a selection of cytochrome P450 enzymes (CYPs 1A2, 2A6, 2B6, 2C19, 2C8, 2C9, 2D6, 2E1, and 3A4).
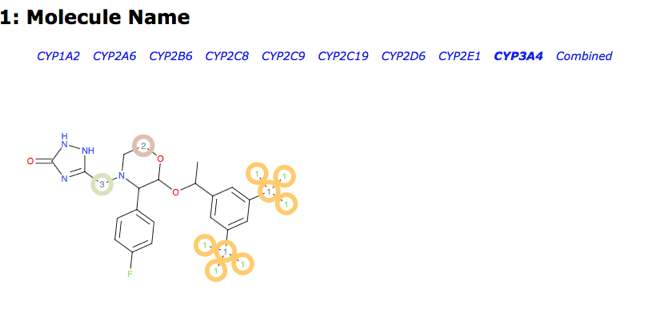
XenoSite is a web-based tool for predicting the atomic sites at which xenobiotics will undergo metabolic modification by Cytochrome P450 enzymes. J. Chem. Inf. Model., 2013, 53 (12), pp 3373–3383 DOI. XenoSite output is interpretable as a probability, which reflects both the confidence of the model that a particular atom is metabolised and the statistical likelihood that its prediction for that atom is correct.
XenoSite combines the computation of multiple quantitative descriptions of molecules, including topological and quantum chemical descriptions, as well as robust descriptions of the reactivity of atomic sites generated by the SmartCyp software [2]. Using a robust neural-network model, XenoSite combines these molecular descriptions with a fingerprint-based search using a technique known as Influence Relevance Voting, a previously developed in silico screening technique. The resulting models of nine CYP isozymes achieve the highest known cross-validated accuracy reported in the literature of CYP metabolism prediction models.
Note, so do not submit confidential structures.
WARNING! XenoSite is provided free-of-charge, however, we cannot make guarantees about data confidentiality on this public website
XenoSite supports models addressing several different enzymes and mechanisms and the results are very nicely displayed on a per model basis (shown using Emend again).
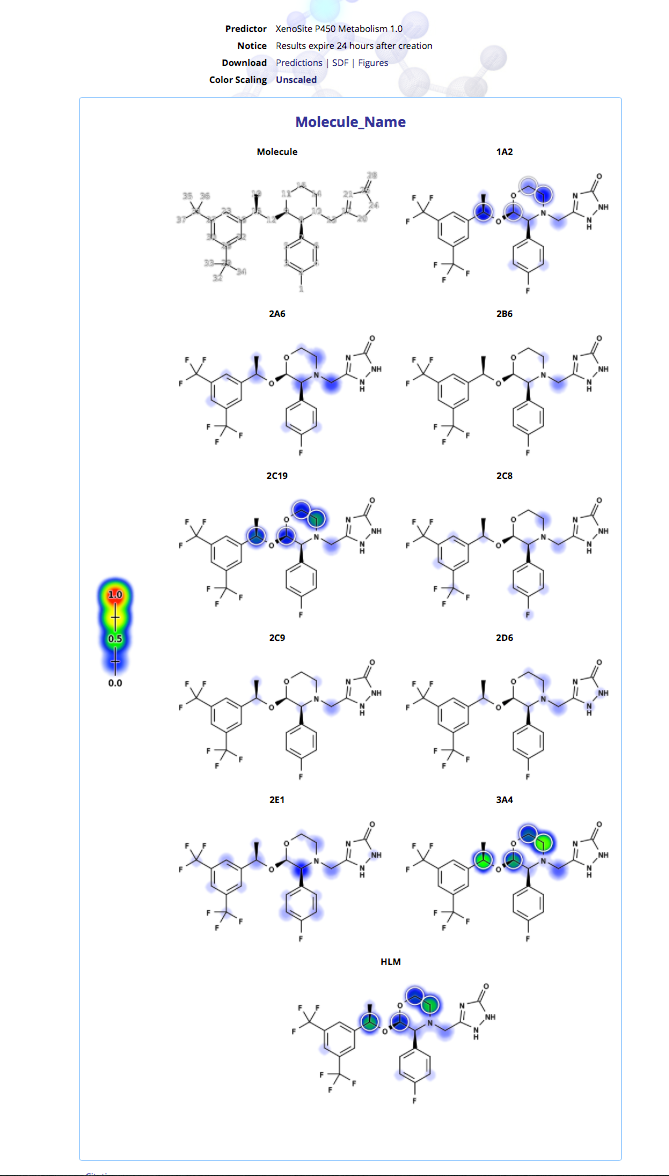
These models can be rather time-consuming and are perhaps best suited for evaluating a few lead compounds.
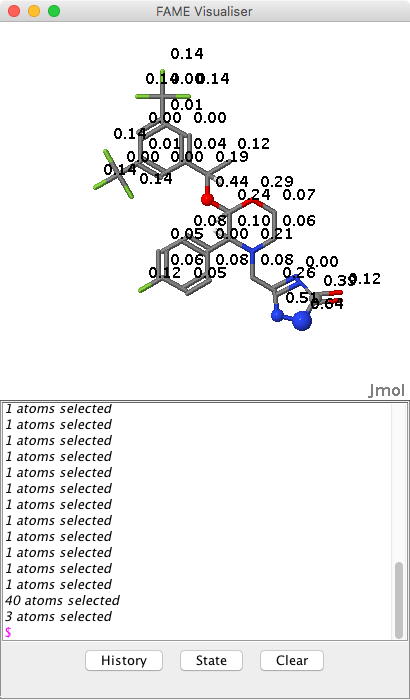
FAME DOI is a collection of random forest models trained on a comprehensive and highly diverse data set of 20,000 small molecules annotated with their experimentally determined sites of metabolism taken from multiple species (rat, dog and human). In addition dedicated models are available to predict sites of metabolism of phase I and II processes. Remarkably this is achieved using only 7 easily calculated descriptors (Table 1), six interpretable atomic descriptors (encoding the element type, hybridization state, and electronic configuration of each atom) and one molecular descriptor (encoding the topological size of a molecule).
In addition dedicated models are available to predict sites of metabolism of phase I and II processes.
FAME 2 DOI builds on this work to improve accuracy, in addition FAME 2 uses a slightly modified version of the visualisation developed by Patrik Rydberg and implemented in SMARTCyp using ChemDoodle Web Components.
It is really useful to have two sites of metabolism tools available that use contrasting methodologies, FAME 2 using curated dataset of experimentally determined metabolism data to build a machine learning model using simple descriptors. In contrast SMARTCyp uses precomputed activation energies from density functional theory (DFT) calculations of model compounds. These are used to predict the reactivity of similar fragments within the target molecule the final score is modified to reflect the accessibility to the active site of the different CYP450 iso forms and improvements for N-oxidations of tertiary amines are included, specifically an empirical corrections to unlikely oxidations of tertiary alkylamines
In FAME 2 rather than using the simple random forest machine learning algorithm used in the original method, an extremely randomised trees approach is used DOI which is a computationally efficient classification algorithm. FAME used a set of 2D descriptors 7 easily calculated descriptors, six interpretable atomic descriptors (encoding the element type, hybridization state, and electronic configuration of each atom) and one molecular descriptor (encoding the topological size of a molecule). In contrast FAME 2 uses circular descriptions of atoms and their environments. As can be seen in the help message below it is possible to change the diameter of the atom encoding fingerprint from 1 to 6. The default 'circCDKATF1' is a model based on the atom itself and its immediate neighbors (atoms at most one bond away).
java -jar /Users/Username/Downloads/fame2/fame2.jar -h
usage: fame2 [-h] [--version] [-m {circCDK_ATF_1,circCDK_4,circCDK_ATF_6}]
[-s [SMILES [SMILES ...]]] [-o OUTPUT_DIRECTORY] [-p] [-c]
[FILE [FILE ...]]
This is fame2. It attempts to predict sites of metabolism for supplied
chemical compounds. It includes extra trees models for regioselectivity
prediction of some cytochrome P450 isoforms.
positional arguments:
FILE One or more SDF files with compounds to predict.
One SDF can contain multiple compounds.
All molecules should be neutral and have explicit
hydrogens added prior to modelling. If there are
still missing hydrogens, the software will try to
add them automatically.Calculating spatial
coordinates of atoms is not necessary.
optional arguments:
-h, --help show this help message and exit
--version Show program version.
-m {circCDK_ATF_1,circCDK_4,circCDK_ATF_6}, --model {circCDK_ATF_1,circCDK_4,circCDK_ATF_6}
Model to use to generate predictions.
Either the model with the best average
performance ('circCDK_ATF_6') during the
independent test set validation as performed in
the original paper or one of the simpler models
that were found to have comparable performance
('circCDK_ATF_1' and 'circCDK_4'). The
'circCDK_ATF_1' model is selected by default as
it is expected to offer the best trade-off
between generalization and accuracy.
The number after the model code indicates how
wide the encodedenvironment of an atom is. For
example, the default 'circCDK_ATF_1' is a model
based on the atom itself and its immediate
neighbors (atoms at most one bond away).
(default: circCDK_ATF_1)
-s [SMILES [SMILES ...]], --smiles [SMILES [SMILES ...]]
One or more SMILES strings of compounds to
predict.
All molecules should be neutral and have explicit
hydrogens added prior to modelling. If there are
still missing hydrogens, the software will try to
add them automatically.
-o OUTPUT_DIRECTORY, --output-directory OUTPUT_DIRECTORY
The path to the output directory. If it doesn't
exist, it will be created. (default: fame_results)
-p, --depict-png Generates depictions of molecules with the
predicted sites highlighted as PNG files in
addition to the HTML output. (default: false)
-c, --output-csv Saves calculated descriptors and predictions to
CSV files. (default: false)
The predictions are generated as a simple HTML page (shown below) which displays the structure of the compound with the predicted SoMs highlighted with yellow circles, moving the cursor over the structure reals the atom numbers that correspond to the numbers in the table.
FAME II Output
Produced: 2017-08-15_20-53-43.
Input file: [/Users/username/Desktop/fame2/example_compounds/tamoxifen.sdf].
Visualization:
To alternate between atoms and atom numbers, move the mouse cursor over the figure.
|
Molecule 2733526
|
StarDrop’s P450 calculations use semi-empirical quantum mechanical calculations (AM1 in MOPAC) to estimate the activation energy for product formation at each potential site of metabolism. These are corrected for steric and orientation effects of the binding pockets of different isoforms, currently covering: CYP3A4, CYP2D6, CYP2C9, CYP2C8, CYP1A2 and CYP2E1. In addition to the regioselectivity of metabolism for each isoform, the models also predict the lability of metabolism of each site in absolute terms and the metabolite formed by metabolism at each site. The calculations run on a server because they are more computationally intensive than QSAR models. They take ~1-2 minutes per ‘drug like’ compound. However they can also be scaled across multiple cores/CPUs to increase throughput.
A comparison of StarDrop and SMARTCyp (where specie isoform models are available).
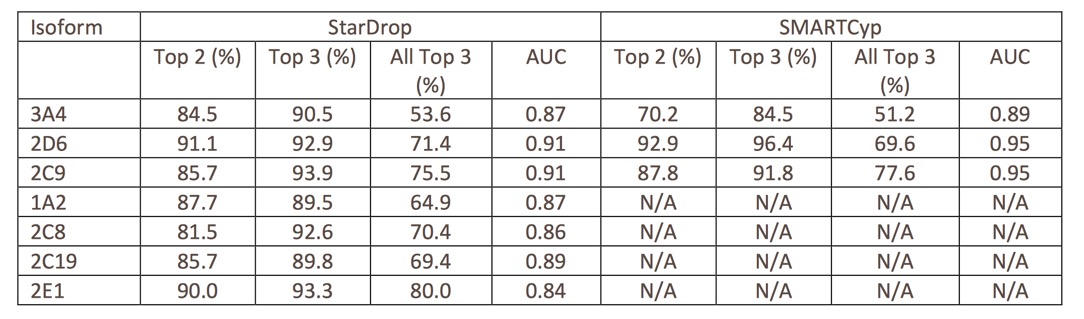
It is noteworthy that SMARTCyp is better at predicting the sites of metabolism for 2D6 and 2C9, this is probably due to more restricted substrate specificity of 2D6 and 2C9. In addition the active site for 3A4 is able to accommodate a much greater diversity of substrates (and poses) meaning that novel substrates outside the applicability domain defined by the fixed list of SMARTS in SMARTCyp may not be predicted. In contrast the semi-empirical quantum mechanical calculations will still be valid.
BioTransformer is a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. BioTransformer combines a machine learning approach with a knowledge-based approach to predict small molecule metabolism in human tissues (e.g. liver tissue), the human gut as well as the environment (soil and water microbiota), via its metabolism prediction tool. In addition BioTransformer provides information on secondary metabolism. There is a review of BioTransformer here
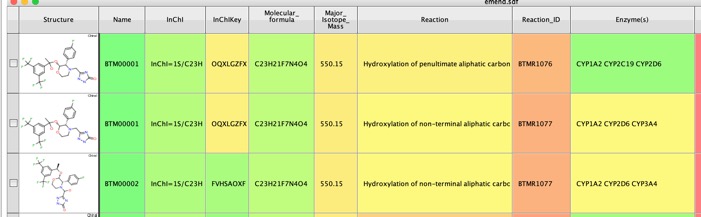
Using Emend as the test molecule BioTransformer identified over 200 putative metabolites. As well as the putative metabolite structure a range of other properties are calculated including InChiKey which can be used to search many other databases, Major Isotopic Mass which could be used in metabolite identification. Together with a description of the reaction and the likely enzymes involved. For the prediction of CYP450 metabolism, BioTransformer makes use of CypReact, a tool for CYP450 substrate specificity prediction
Running the process of Diazepam yielded a sdf file containing yielded 151 structures but on closer examination there were many duplicate structures and only 51 unique structures (I used this Vortex script to flag duplicate structures). This is because several reaction patterns can yield the same structure. Around 30 had PubChem CID suggesting these are previously identified metabolites.
Phase II metabolism
Aspirin hydrolyses to produce salicylic acid (2-hydroxybenzoic acid), Phase II metabolism results in conjugates with glycine (to form salicyluric acid) or glucuronic acid to make several ionised metabolites that can then be excreted in the urine.
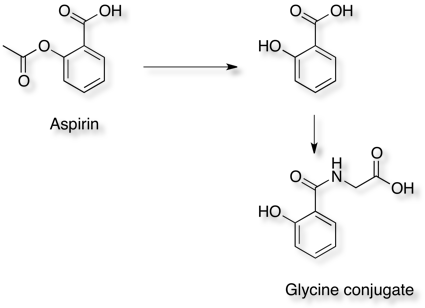
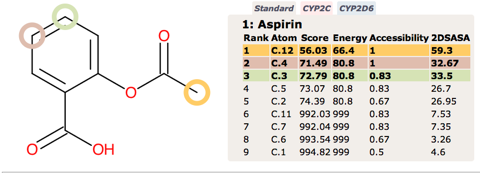
Since none of the processes described above are CYP mediated it is not surprising that they are not highlighted by SMARTCyp.
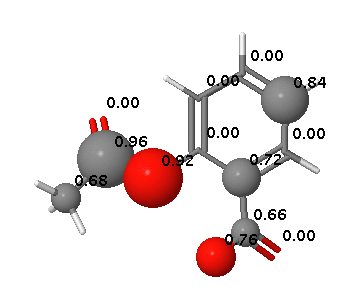
In contrast FAME correctly identifies hydrolysis of the ester and the potential of the carboxylic acid to be involved in metabolic processes.
FAME offers a high performance prediction of sites of metabolism mediated by a wide variety of mechanisms.
GLORYx DOI predicts phase I and phase II metabolites for the chemical compound(s) provided by the user. The method is based on the FAME site of metabolism (SoM) prediction combined with sets of reaction rules encoding both phase I and phase II metabolic reactions. GLORYx is freely available as a web server at https://nerdd.zbh.uni-hamburg.de/ and is available as a java jar file upon request.
The GLORYx can be run from the command line. All you need to do is specify one or more input molecules, as SMILES strings or as an SDF file.
java -Xmx16g -jar /Users/chrisswain/Desktop/gloryx_2020-09-02/gloryx.jar -s "FC(F)(F)c1cc(cc(c1)C(F)(F)F)[C@H](O[C@H]4OCCN(CC/2=N/C(=O)NN\2)[C@H]4c3ccc(F)cc3)C"
Note that since the FAME 3 models can take quite a bit of memory, the -Xmx16g flag may be necessary. On my MacPro it utilised multiple cores
The metabolism phase can be specified. For example, to predict metabolites for only phase I
java -Xmx16g -jar /Users/chrisswain/Desktop/gloryx_2020-09-02/gloryx.jar -s "FC(F)(F)c1cc(cc(c1)C(F)(F)F)[C@H](O[C@H]4OCCN(CC/2=N/C(=O)NN\2)[C@H]4c3ccc(F)cc3)C" -p P1
The output is stored in a folder entitled "metabolitepredictionresults" created in the users home folder. This folder will contain the predictions in the form of one or more SDF files, whereby each SDF file corresponds to up to 1000 input molecules. The input molecules are included as the first record in the output file(s), so that each input molecule is followed by its predicted metabolites.
The resulting output file contains a large number of predicted metabolites together with a score, a description of the type of reaction and the InChi. This looks to be really comprehensive and would be very useful for those involved in metabolite ID.
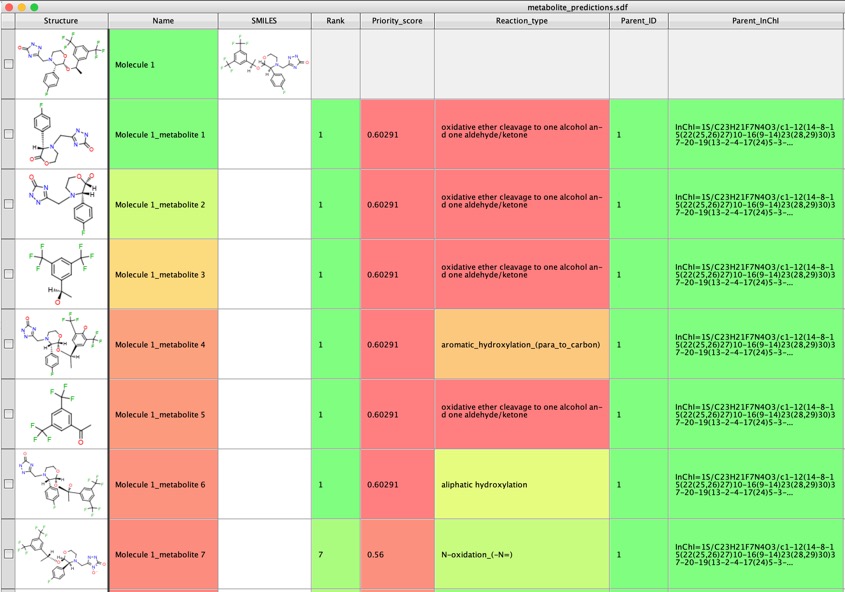
Running the prediction with aspirin as the input highlights a variety of non-CYP mediated metabolic pathways.
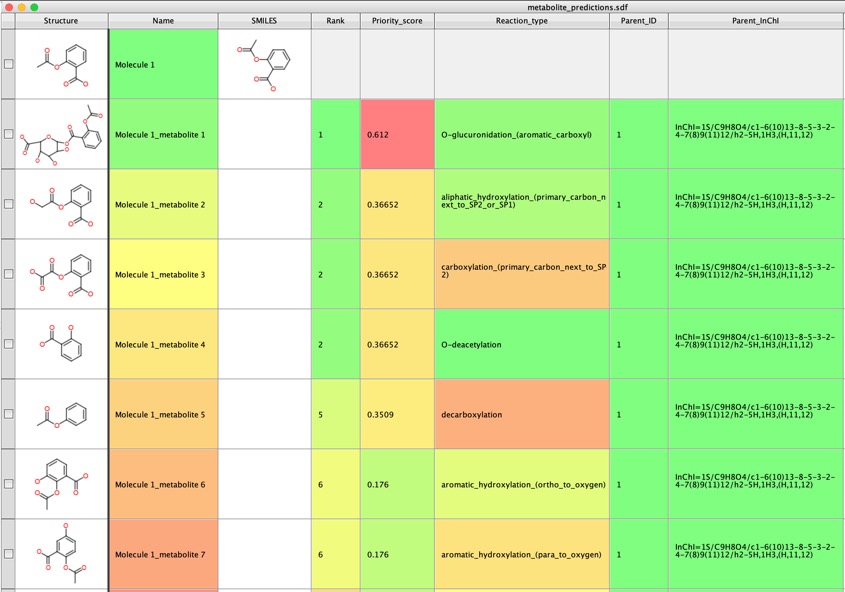
I tried a range of other molecules and GLORYx was really very impressive in identifying potential metabolites.
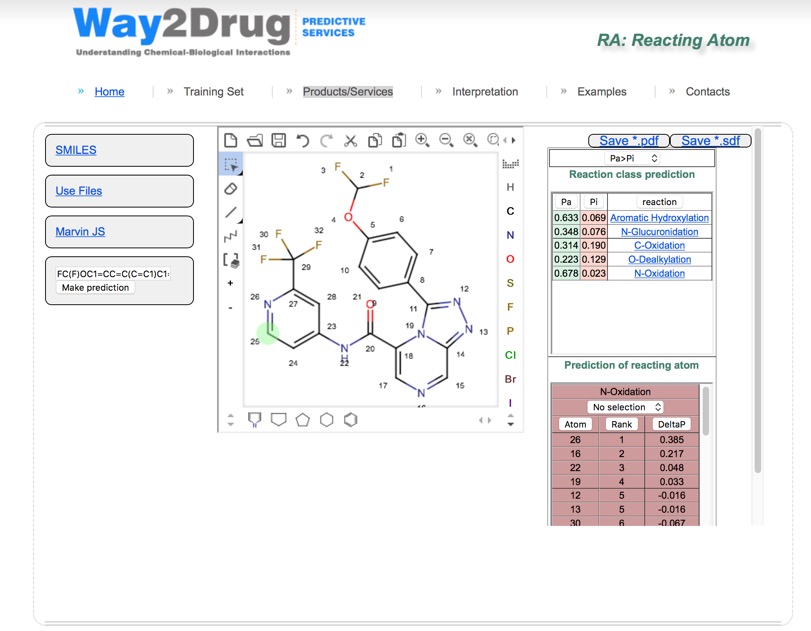
Way2Drug offers a web service for predicting sites of metabolism details of which have been published DOI. However I would not recommend that you use it for proprietary molecules. All major classes of metabolic reactions—aliphatic and aromatic hydroxylation, N- and O-glucuronidation, N-, S- and C-oxidation, and N- and O-dealkylation are evaluated.
Global solutions, intended to predict the metabolism of any molecule exposed to a complex biological system. This type of solution is often rule-based and uses an extensive database of known biotransformations Programs such as MetaDrug (Expert Opin. Drug Metab. Toxicol. (2005) 1(1)) uses a series of rules together with a series of QSAR models to predict metabolic transformations, and includes both phase I and phase II metabolism. The transformations described include among many:- C,N,S and P-oxidation, including dealkylation, hydroxylation, double bond peroxidation, Quinone formation, reduction (nitro, carbonyl, azo, sulphur), hydrolysis (esters, amides, phosphates, epoxides), glucoronidation, sulphation, glutathione conjugation, methyl transferases, amino acid conjugation. Other programs using similar approaches include Meteor, (Pure Appl.Chem., Vol.76, No.5, pp.907–914, 2004) and Meta.
In general I’ve found these products very useful for identifying all potential metabolic sites, however they can over-predict and you may well find many of the potential metabolic routes have negligible contributions in vivo.
Whilst the above programs offer a nice insight into the potential soft spots in the molecule sometimes you just want to know which enzymes are likely to be involved in the metabolism of a molecule, CypReact DOI takes a structure (SMILES or sdf input) and predicts if the molecule will react with any one of the nine of the most important human cytochrome P450 (CYP450) enzymes [CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, or CYP3A4]. CypReact is a command line tool, is extremely fast and is ideal for quickly evaluating a batch of compounds. CypReact is available at https://bitbucket.org/Leon_Ti/cypreact.
If we look at Emend
MacPro:~ Chris$ java -jar /Users/Chris/Downloads/Leon_Ti-cypreact-29a582219630/CypReactBundle/cypreact.jar /Users/Chris/Downloads/Leon_Ti-cypreact-29a582219630/CypReactBundle/ /Users/Chris/Desktop/SampleFiles/emend.sdf /Users/Chris/Desktop/SampleFiles/emendmetab.sdf 1A2,2A6,2B6,2C8,2C9,2C19,2D6,2E1,3A4
Processing Molecue: 1
--------------------------------------
1A2, Mole1: R
--------------------------------------
3A4, Mole1: R
--------------------------------------
2B6, Mole1: N
--------------------------------------
2E1, Mole1: N
--------------------------------------
2C9, Mole1: R
--------------------------------------
2C19, Mole1: R
--------------------------------------
2D6, Mole1: R
--------------------------------------
2C8, Mole1: N
--------------------------------------
2A6, Mole1: N
IMPACTS
IMPACTS (In-silico Metabolism Prediction by Activated Cytochromes and Transition States) is a hybrid site-of-metabolism (SoM) identification tool which combines docking to CYP enzymes, transition state (TS) modeling, and rule-based substrate reactivity prediction to predict the SoM of xenobiotics. The input is a 3D structure with the correct protonation state, Molecular Forecaster have their own drawing tools but since the file format is .mol2 these could be generated using a variety of alternative tools. The output includes the structure bound to the CYP and the putative metabolites.
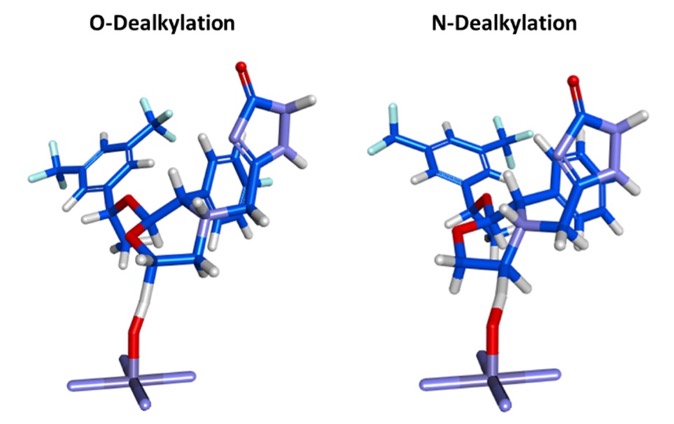
Campagna-Slater V., Pottel J., Therrien E., Cantin L.-D., Moitessier N., J Chem Inf Model, 2012, 52, 2471-2483 DOI
Worth reading:
Computational Prediction of Metabolism: Sites, Products, SAR, P450 Enzyme Dynamics, and Mechanisms DOI
Predicting Regioselectivity and Lability of Cytochrome P450 Metabolism Using Quantum Mechanical Simulations DOI
See also the section on Metabolism
Updated 8 December 2022