Cytochrome 3A4 inhibition
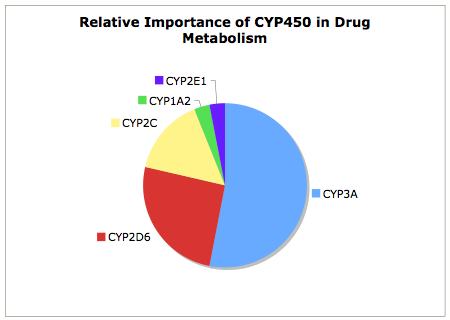
It is useful to remember of the 57 Human CYP Enzymes few have major role in drug metabolism, as can be seen from the chart below CYP3A is by far the major contributor to CYP450 mediated metabolism, this together with CYP2D6 accounts for over 75% of the CYP450 mediated metabolism.
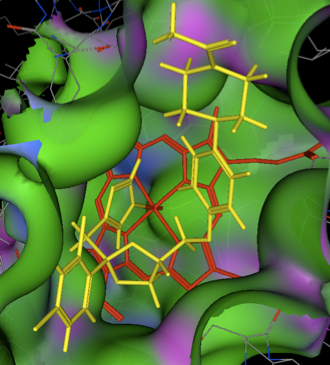
Active site of CYP3A4
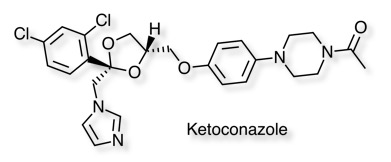
The large lipophilic binding site of CYP3A4 can accommodate a wide variety of inhibitors. A crystal structure of Ketoconazole bound to human CYP3A4 is available, this is the crystal structure 2V0M displayed in MOE, ketoconazole is shown in yellow and the haem in red. The nitrogen of the imidazole is bound to the iron, the surface of the protein is coloured coded green= lipophilic, purple= polar. The majority of the binding site is open and lipophilic and even a relatively large molecule like ketoconazole is comfortably accommodated. Other than the coordination to the iron the other major interactions are arene-arene stacking.
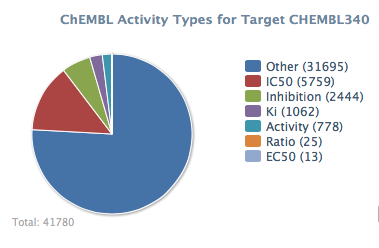
CYP3A4 Inhibition Data from ChEMBL
ChEMBL is a database of bioactive drug-like small molecules, it contains 2-D structures, calculated properties (e.g. logP, Molecular Weight, Lipinski Parameters, etc.) and abstracted bioactivities (e.g. binding constants, pharmacology and ADMET data). Currently it contains data for 1,359,508 molecules and 9,414 different targets. ChEMBL contains data for CYP3A4 inhibitors, looking through the data the largest activity type is “other” this seems to contain a wide variety of different experimental data (off rate, time dependent inhibition, selectivity ratio over other target etc.), the next largest is IC50 data, “Inhibition” contains % inhibition at various concentrations.
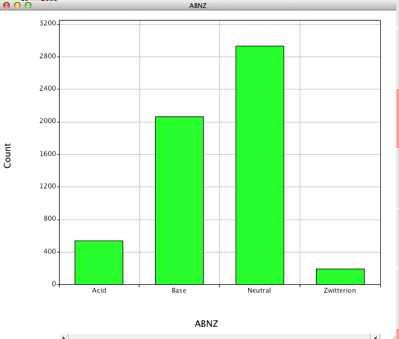
I downloaded all the IC50 data and imported it into Vortex, I then deleted those results that the ChEMBL team had flagged as potentially unreliable. Then I calculated a number of physicochemical properties using a variety of scripts. Looking at the potential ionisable groups it is clear that the majority of the molecules are neutral or basic with significantly less acids or zwitterions.
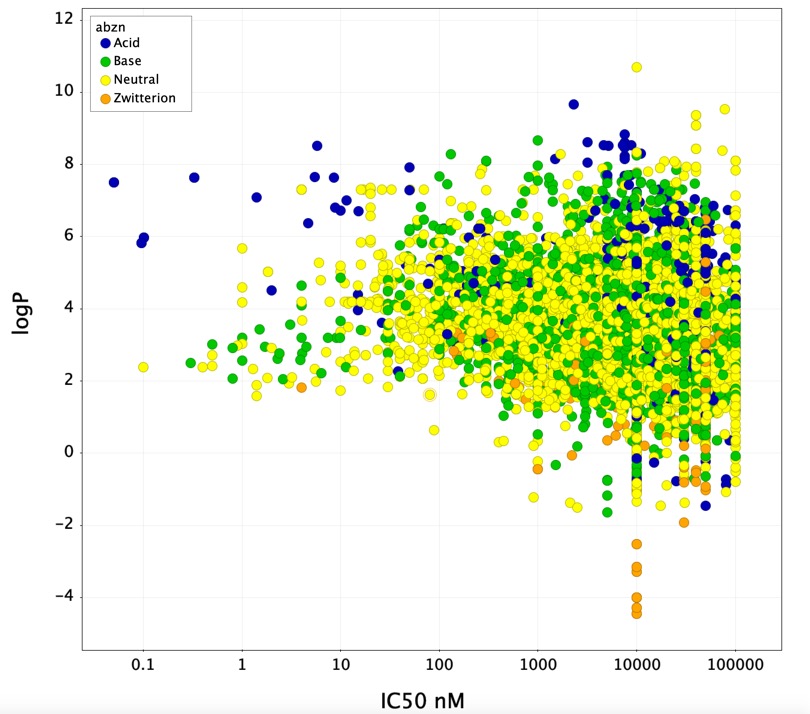
Looking at the scatter plot below (IC50 v calc LogP) it is noticeable that in general zwitterions do not inhibit CYP3A4, in addition few acids display high affinity except for a group of very lipophilic compounds. Indeed there are several examples where introduction of a carboxylic acid reduces CYP3A4 inhibition. Usually lipophilicity is thought to have a role in determining CYP inhibition, however the CYP3A4 data from ChEMBL would suggest that this is not necessarily the case.
Matched Pair Analysis
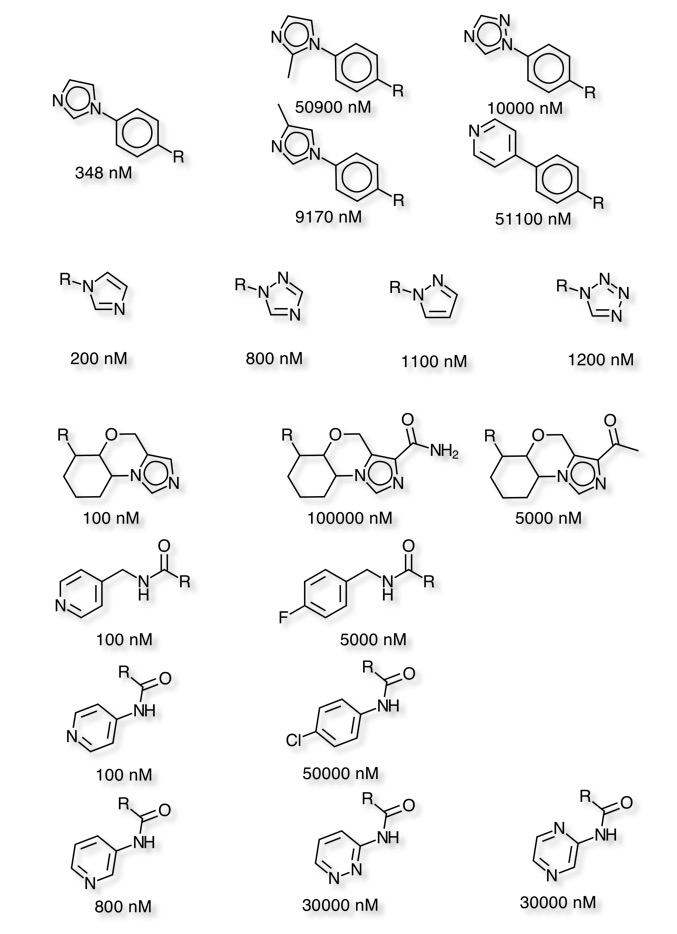
Using Vortex to conduct a matched pair analysis helps to identify small changes that have a significant influence on the CYP3A4 inhibition. As might be expected many of the more potent inhibitors have a heterocycle that can coordinate to the iron in the haem ring of the cytochrome. The matched pair analysis identifies a number of examples where modification of the heterocycle has a significant effect on inhibition. Either by sterically encumbering the nitrogen proposed to coordinate to the iron, or by reducing the electron density on the nitrogen. An alternative approach has been to replace a pyridine with the bioisosteric fluoro or chloro phenyl
See Also
Last Updated 11 November 2018