Aldehyde Oxidase
I first became aware of Aldehyde Oxidase when some of my co-workers (http://dx.doi.org/10.1080/00498250600567903) highlighted the metabolism of a functionally selective, inverse agonist at the benzodiazepine site of GABAA alpha5 containing receptors. During the ADME studies it was observed that one metabolite was absent in Dog.
Subsequent in vitro and in vivo studies demonstrated that the metabolic behaviour was consistent with that previously reported for aldehyde oxidase substrates, in that the aldehyde oxidase metabolites are formed in rat, rhesus monkey and human, but not in dog. I was reminded of this work when I recently saw an excellent presentation by Drew Gibson (Pfizer).
Aldehyde oxidase AOX1 (EC1.2.3.1) is a non-NADPH dependent, molybdenum cofactor containing soluble enzyme present in the liver and other tissues of several mammalian species, and whilst Cytochrome P450 enzymes have been extensively studied and for part of most drug discovery cascades it worth remembering they are not the only sources of xenobiotic metabolism. Aldehyde oxidase might be of particular interest for certain chemotypes, because in addition to the oxidation of aldehydes to carboxylic acids it is also responsible for the oxidation of nitrogen-containing heterocyclic systems such as pyridines, diazines, benzimidazoles, purines and a wide variety of other fused heteroaromatic systems. Drug Metab. Pharmacokinet. 2006, 21, 83–98. Oxidation by AO is thought to proceed by nucleophilic attack of a high-valent molybdenum-oxo species on aromatic carbon atoms adjacent to nitrogen.
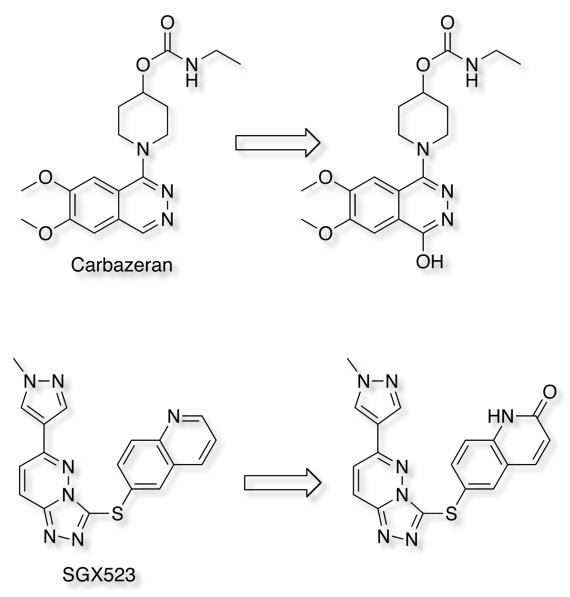
Carbazeran failed in clinical studies due to lack of exposure after oral dosing, later shown to be due to extensive hydroxylation mediated by AOX1 metabolism, this was not apparent in preclinical studies in dog.
The issue of species differences has been investigated in some detail (doi:10.1074/jbc.M600850200) see table below. Rodents synthesise by four aldehyde oxidases AOX1 and aldehyde oxidase homologs 1-3 (AOH1, AOH2, and AOH3) in contrast Humans synthesise a single functional aldehyde oxidase, AOX1. The dog genome contains two structurally conserved and active aldehyde oxidases clustering on chromosome 37. These have been shown, by cloning of the corresponding cDNAs, that they are the orthologs of rodent AOH2 and AOH3. Monkey appears to be very similar to human. Human aldehyde oxidase 1 (AOX1) has been subcloned (doi: 10.1124/dmd.109.029520) into a vector suitable for expression in Escherichia coli, and the protein has been expressed. The resulting protein is active, with sulfur being incorporated in the molybdopterin cofactor. Aldehyde Oxidase is a cytosolic enzyme and so most liver microsomal assays may under estimate the contribution AO may make towards clearance of a compound, this serves to emphasise the importance of hepatocyte based assays that containing all the metabolising machinery. The S9 fraction from human liver microsome preparations will also contain cytosolic enzymes.
| Species | AOX1 | AOX2 | AOX3 | AOX4 |
|---|---|---|---|---|
| Human | Yes | No | No | No |
| Dog | No | Yes | No | Yes |
| Rat | Yes | Yes | Yes | Yes |
| Mouse | Yes | Yes | Yes | Yes |
| Rabbit | Yes | Yes | Yes | Yes |
| Rhesus | Yes | Yes | No | Yes |
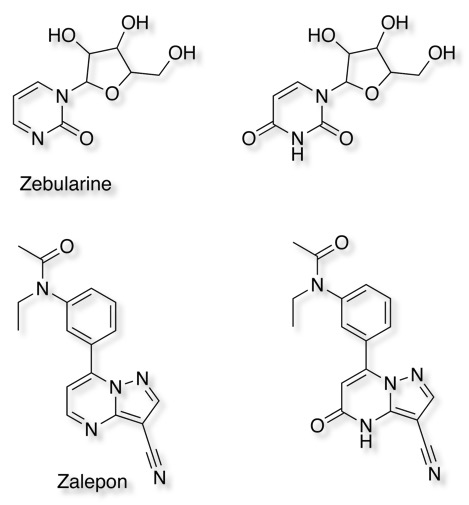
More recently the development of the Selective MET receptor tyrosine kinase SGX523 was halted due to AOX1 mediated metabolism seen in human studies but also renal toxicity that might be caused by the very poor solubility of the metabolite (doi:10.1124/dmd.110.032375). Other examples of AOX1 metabolism include Zebularine (doi:10.1016/j.bmc.2005.07.053 ), Zalepon (xenobiotica, 2002, vol. 32, no.10, 835-847 ).
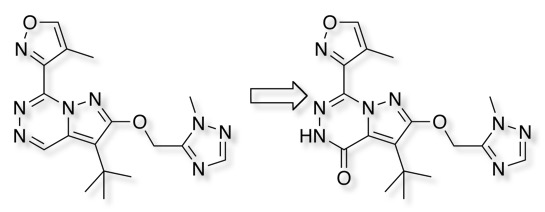
Given the current interest in kinase inhibitors, many of which have nitrogen containing heterocycles as the core structure, it has been suggested that Aldehyde Oxidase be included in the ADME/T screen.
Predicting potential sites of metabolism
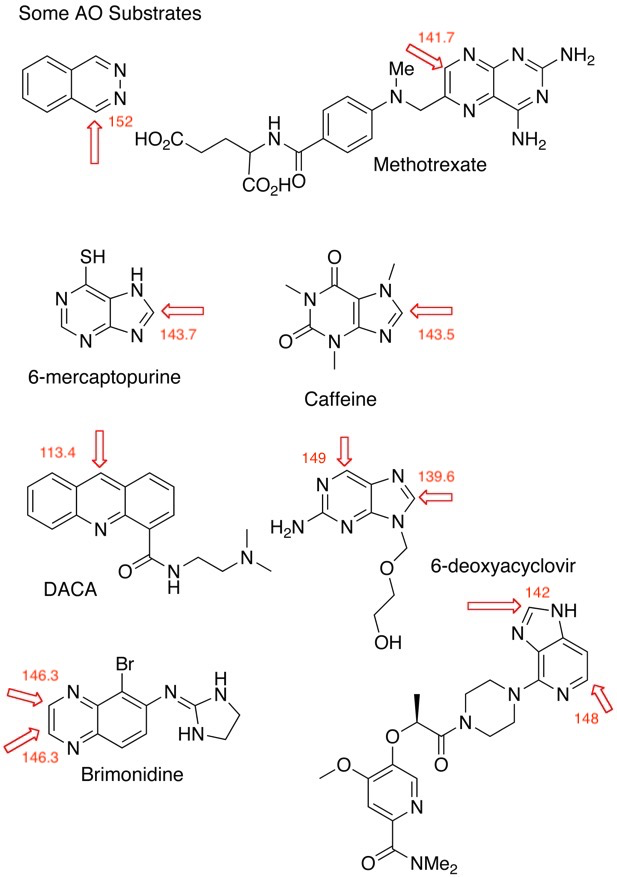
Oxidations usually occur at the electron-deficient sp2 carbons adjacent to nitrogen and studies have shown that the potential sites of metabolism can be identified by calculating point charges using DFT methods DOI, in addition 13C shifts has been shown to be a reasonable surrogate. While this can be measured experimentally for known compounds, for prediction in silico for proposed structures NMR prediction software may also be useful. I've annotated some known AO substrates (below) with 13C shifts calculated using https://nmrshiftdb.nmr.uni-koeln.de/. In the majority of cases a trend of greater AO susceptibility where shifts of sp2 carbons are greater than 140 ppm is observed.
The table below highlights some of the issues that have been observed in clinical development for a range of compounds, including high metabolic clearance, renal toxicity due to insoluble metabolites and bioactivation.
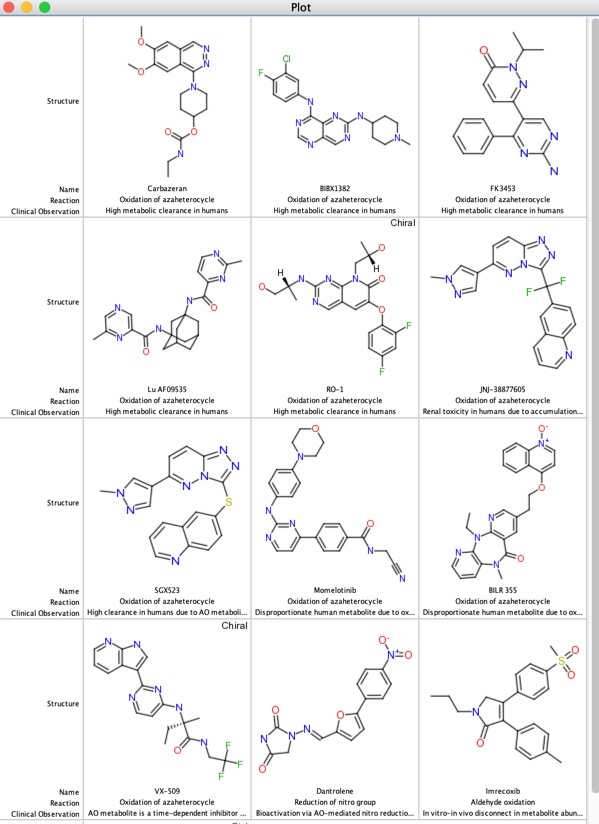
AO substrate activity does not appear to correlate with physicochemical properties, but the presence of a N-heterocycle is probably a useful flag. Potential solutions include blocking the site of metabolism or using an alternative heterocycle. Aldehyde Oxidase has also been shown to be responsible for the reduction on some functional groups including N-oxides, nitro, sulphoxide and some heterocycles.
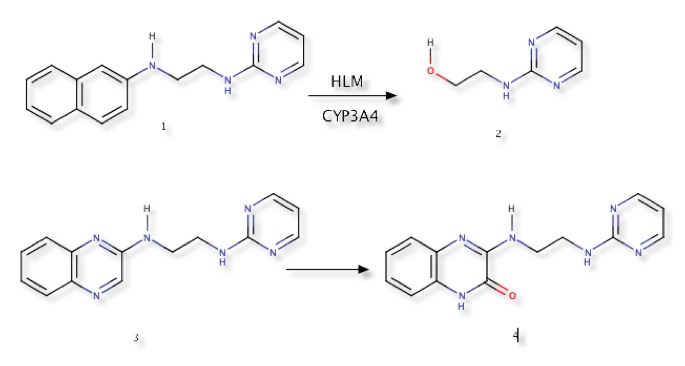
A salutary lesson has been described DOI in which there was a metabolic shift from CYP mediated oxidation to AOX mediated metabolism during the course of a MedChem program. When compound 1 (below) was incubated in human liver microsomes (HLM), it demonstrated notable metabolic instability. After 60 minutes the compound was totally degraded into three metabolites, including 2, Incubation with recombinant CYP isoforms demonstrated the preeminent role of the enzyme CYP-3A4. In an effort to improve metabolic stability the naphthyl group was replaced by the less election-rich quinoxalin group (3), this proved to be stable in human liver microsomes. However subsequent experiments showed the compound was unstable in human liver cytosol. The effort to protect the molecule from cytochrome P450 metabolism had inadvertently shifted the metabolism to AOX.
Inhibitors
The inhibition of Aldehyde Oxidase by 239 known drugs has also been published DOI: 10.1177/0091270003260336 a number of compounds displayed sub-micromolar activity.
Drug IC50 (µM)
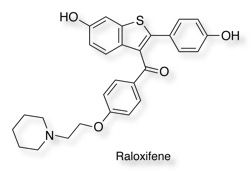
Raloxifene 0.0029
Perphenazine 0.033
Thioridazine 0.16
Menadione 0.20
Trifluperazine 0.24
Amitriptyline 0.26
Estradiol 0.29
Felodipine 0.30
Clomipramine 0.48
Loratidine 0.49
Promethazine 0.51
Chlorpromazine 0.57
Ethinyl estradiol 0.57
Norclomipramine 0.60
Amodiaquine 0.74
Nortriptyline 0.85
The inhibition of AOX1 by Raloxifene has been studies in some detail. At least one of the two phenolic moieties is essential for raloxifene to exhibit its potent inhibition, the dimethoxy derivative being 1000-fold less potent. The hydrophobic 2-piperidinylphenoxyethyl portion was also needed for inhibition; however, the basicity of the piperidine nitrogen was not essential.
Structure of AOX1 and Mechanism of oxidation
In the first step, the site of oxidation, (he electrophilic carbon of the azaheterocycle or aldehyde), is attacked by the nucleophilic -OH ligand of the MoCo ligand (MoVI), this is likely further activated by Glu1270 (shown in blue below), hydride transfer to MoCo’s sulfido ligand occurs simultaneously (via a tetrahedral intermediate) to form a transition state stabilised by hydrogen bonding from Glu1270 and Lys893. This nucleophilic attack is in contrast to the electrophilic oxygen mechanism of CYP450 oxidation, leading to different sites of metabolism.
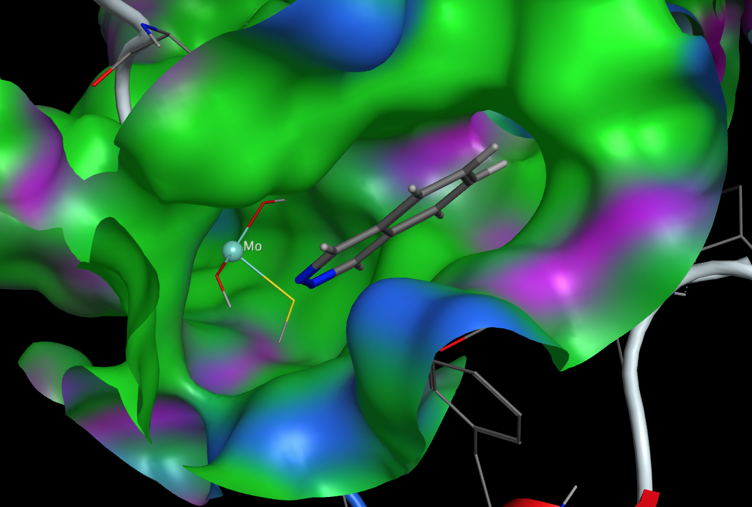
Recently the crystal structures of human AOX1, in its substrate-free form PDB ID: 4UHW, as well as in complex with both the substrate, phthalazine (red, space filling), and the inhibitor, thioridazine (yellow, stick) PDB ID: 4UHX, have been reported DOI, the molybdenum cofactor is shown in green.
Mouse Controls
| Movement | Mouse Input | Touch Input | ||
|---|---|---|---|---|
| Rotation | Primary Mouse Button | Single touch | ||
| Translation | Middle Mouse Button or Ctrl+Primary | Triple touch | ||
| Zoom | Scroll Wheel or Second Mouse Button or Shift+Primary | Pinch (double touch) | ||
| Slab | Ctrl+Second | Not Available |
A closer examination of the active site below (Created using MOE, green = lipophilic, blue polar) shows a flattened lipophilic pocket ideal for accommodating aromatic systems.
A simple test
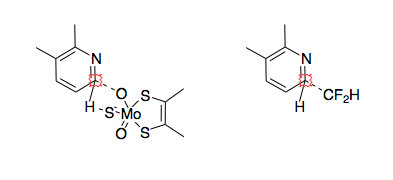
A recent publication DOI suggests a simple test for the early identification of heteroaromatic drug candidates that have a high probability of metabolism by AO. Bis(((difluoromethyl)sulfinyl)oxy)zinc (DFMS) was used as a source of the CF2H racial, simple LCMS was then used to identify a characteristic M+50 peak. It is also possible to scale up and isolate these metabolically blocked compounds and retest them for improved qualities.
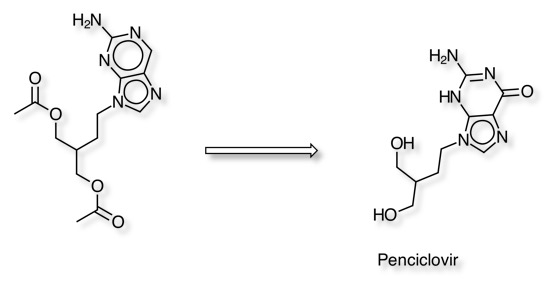
Prodrug Activation
Penciclovir is an antiviral drug with very poor bioavailability (F=1.5%), Famciclovir is a prodrug of Penciclovir and has much improved bioavailability up to 77% DOI. Bioactivation is in part by aldehyde oxidase.
Approaches to mitigate AO mediated metabolism
Given the diversity of substrates it is perhaps not surprising that most computational moulds can be used to predict potential sites of metabolism but this really needs to be checked experimentally. If AO-catalyzed oxidations of the chemotype are confirmed, medicinal chemists have a couple of options provided the exact site of metabolism can be conformed. You can proceed at risk and try to predict human clearance based on preclinical models. Alternatively, it might be block the site of metabolism or use ring substitution to alter the electrostatics or sterics and thus reduce the rate of metabolism. Typical modifications may include blocking the site of metabolism, steric substitution next to the site, changing ring size, saturating the ring, or modulating the electrostatics.
Worth reading
Aldehyde Oxidase: An Enzyme of Emerging Importance in Drug Discovery, J. Med. Chem. 2010, 53, 8441–8460 DOI
A Simple Litmus Test for Aldehyde Oxidase Metabolism of Heteroarenes J. Med. Chem. DOI
Structure and function of mammalian aldehyde oxidases Arch Toxicol (2016) 90:753–780. DOI
Metabolism by aldehyde oxidase: drug design and complementary approaches to challenges in drug discovery. DOI.
Updated 7 December 2019