Target Validation
A couple of minor updates to the Target Validation page.
https://www.cambridgemedchemconsulting.com/resources/targetvalidation.html
Reproducibility Project: Cancer Biology
I've been waiting for this for a while. Reproducibility Project: Cancer Biology
The Reproducibility Project: Cancer Biology was an 8-year effort to replicate experiments from high-impact cancer biology papers published between 2010 and 2012. The project was a collaboration between the Center of Open Science and Science Exchange with all papers published as part of this project available in a collection at eLife and all replication data, code, and digital materials for the project available in a collection on OSF.
The work tried to repeat 193 experiments from 53 papers and found a significant number of challenges.

In summary
- Replication effect sizes were 85% smaller on average than the original findings
- 46% of effects replicated successfully on more criteria than they failed
- Original positive results were half as likely to replicate successfully (40%) than original null results (80%)
This quote from In the Pipeline is perhaps a useful reminder.
A robust result can probably be reproduced even if you switch to a different buffer, or if your cell lines have been passaged a different number of times, or if the concentration of the test molecule is a bit off, etc. The more persnickity and local the conditions have to be, the less robust your result is, and in general (sad to say) the lower the odds of it having a real-world impact in drug discovery. There are certainly important things that can only be demonstrated under very precise conditions, don’t get me wrong – but when you’re expecting umpteen thousand patients to take your drug candidate and show real effects, your underlying hypothesis needs to be able to take a good kicking and still come through.
Alphafold in Opentargets
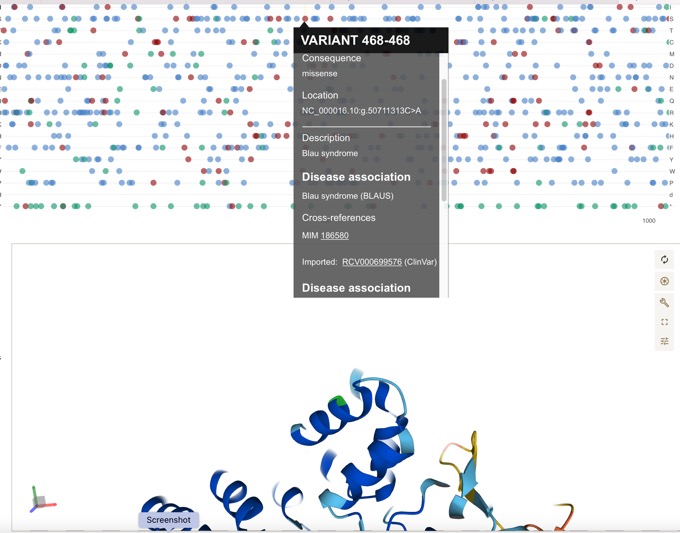
Thanks to fantastic work from the folks at UniProt, the Open Targets Platform target profile pages now feature DeepMind’s AlphaFold data.
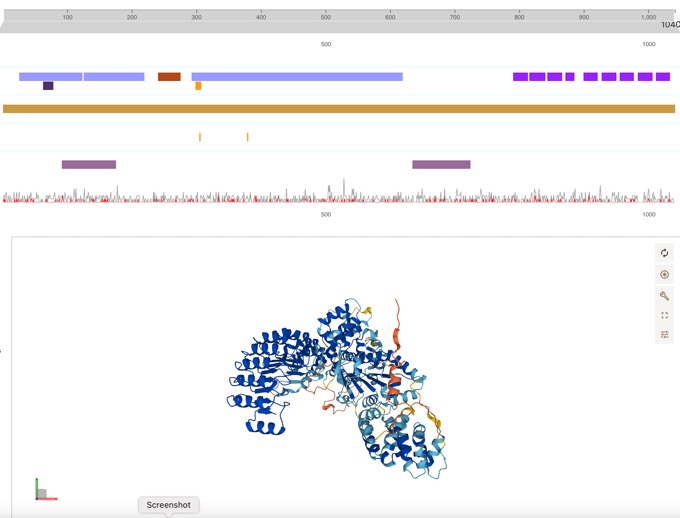
Can be easily linked to disease associations.
Open Targets Platform 21.06 has been released!
Open Targets Platform 21.06 has been released
The Open Targets Platform is a comprehensive tool that supports systematic identification and prioritisation of potential therapeutic drug targets. By integrating publicly available datasets including data generated by the Open Targets consortium, the Platform builds and scores target-disease associations to assist in drug target identification and prioritisation. It also integrates relevant annotation information about targets, diseases, phenotypes, and drugs, as well as their most relevant relationships.
Currently there are:-
Targets 60,606
Diseases 18,507
Drugs 13,185
Evidence strings 13,267,236
Associations 11,755,362
Open Targets Platform: rebuilt, redesigned, reimagined
The Open Targets Platform has been updated.
- The new design of the Platform improves user experience and access to information. It has been reimagined to facilitate the building of new therapeutic hypotheses;
- New features include data on binary molecular interactions, and black box warnings;
- A complete refactoring of the codebase enables rapid development and new deployment strategies.
Chemical Probes
A really interesting review of the Chemical Probes portal
2020 was the first year of visible activity on the Chemical Probes Portal since 2017, with 115 probes added and over 500 compounds now included on the Portal. To celebrate, we’re highlighting ten of the best probes added to the Portal and evaluated by our Scientific Advisory Board in 2020. These probes are selective, potent, cell-active molecules that are rated four stars for use in cells and target new proteins or have new mechanisms of action. They include probes for previously ‘undruggable’ cancer targets, compounds that target GPCRs, epigenetic modulators and PROTACs.
Full details are here https://www.chemicalprobes.org/news/2020s-top-probes.
Open Targets Platform beta test
If you have a little time why not drop by the Open Targets website and give the beta test version and give them some feedback. Full details are in the blog post.
The beta version makes the most of the data from the recent 21.02 release and you can also interrogate the data using the brand new GraphQL API.
In particular, this version features:
- Redesigned evidence pages
- Updated drug profile pages
- More complete disease profile pages
Open Targets is a public-private partnership that uses human genetics and genomics data for systematic drug target identification and prioritisation. The current focus is on oncology, immunology and neurodegeneration.
Generating and interpreting the data required to identify a good drug target demands a diverse set of skills, backgrounds, evidence types and technologies, which do not exist today in any single entity. Open Targets brings together expertise from seven complementary institutions to systematically identify and prioritise targets from which safe and effective medicines can be developed.
Open Targets Platform updated
Target Validation is the most critical step in the Drug Discovery process, almost everything else we can fix. Which is why the update to the Open Targets Platform is so valuable.
The latest release of the Open Targets Platform - 20.09 - is now available at https://www.targetvalidation.org/.
This release sees the addition of ClinGen to the expert curated evidence sources for rare disease genetics that includes UniProt, the Genomics England PanelApp, and Gene2Phenotype.
This latest update includes 27,610 Targets, 13,944 Diseases, 8,419,186 Evidence and 6,551,303 Associations
OpenTargets updated
Just had an email about the latest Open Targets Platform release - 19.11.
In this release there is data on
- 27,069 targets
- 13,579 diseases
- 8.91 million pieces of evidence
- 6.33 million associations between targets and diseases
History of rare diseases and their genetic causes - a data driven approach
One of the advantages of being a consultant is that I can feel free to contribute to projects that I find interesting. So as well as working with a couple of Open-Source drug discovery projects (e.g. Open Source Antibiotics I can also follow a couple of rare disease programs.
This publication looks very useful History of rare diseases and their genetic causes - a data driven approach.
This dataset provides information about monogenic, rare diseases with a known genetic cause supplemented with manually extracted provenance of both the disease and the discovery of the underlying genetic cause of the disease.
More details of how the dataset was constructed.
We collected 4166 rare monogenic diseases according to their OMIM identifier, linked them to 3163 causative genes which are annotated with Ensembl identifiers and HGNC symbols. The PubMed identifier of the scientific publication, which for the first time describes the rare disease, and the publication which found the gene causing this disease were added using information from OMIM, Wikipedia, Google Scholar, Whonamedit, and PubMed. The data is available as a spreadsheet and as RDF in a semantic model modified from DisGeNET.
A very interesting read.
Open Targets Updated
The latest Open Targets Platform 19.09 has been released. The latest release contains
27,024 targets 10,474 diseases 3.33 million pieces of evidence 7.78 million associations between targets and diseases
In addition, a number of Target Enabling Packages (TEP) provided by Structural Genomics Consortium have been included, there are more details here. Several new chemical probes have also been included.
canSAR BLACK
cansar black v1.1.1 now available - includes improved search, new protein family page, and performance improvements
canSAR is an integrated knowledge-base that brings together multidisciplinary data across biology, chemistry, pharmacology, structural biology, cellular networks and clinical annotations, and applies machine learning approaches to provide drug-discovery useful predictions. canSAR’s goal is to enable cancer translational research and drug discovery through providing this knowledge to researchers from across different disciplines. It provides a single information portal to answer complex multi-disciplinary questions including - among many others: what is known about a protein, in which cancers is it expressed or mutated and what chemical tools and cell line models can be used to experimentally probe its activity? What is known about a drug, its cellular sensitivity profile and what proteins is it known to bind that may explain unusual bioactivity?
Open Targets Platform: release 19.06 is out
The latest update of the Open Targets Platform, release 19.06 is available.
This update includes
Target safety information
As a follow-up to the safety data in Open Targets Platform release 19.04, now has more targets with known safety effects and safety risk information, including TBXA2R and JAK2.
TEPs and chemical probes
In this release, they've included the latest Target Enabling Packages (TEPs) for GALT, GALK1 and MLLT1. Also added more chemical probes, small-molecule modulators of a protein’s function that can be used in cell-based or animal studies.
Target-disease associations
A new release always means new evidence available for novel target-disease associations.
No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples
For depression, SLC6A4 seemed like a great candidate and was supported by very early gene studies
BUT….
Am J Psychiatry. 2019 May 1;176(5):376-387. DOI
The study results do not support previous depression candidate gene findings, in which large genetic effects are frequently reported in samples orders of magnitude smaller than those examined here. Instead, the results suggest that early hypotheses about depression candidate genes were incorrect and that the large number of associations reported in the depression candidate gene literature are likely to be false positives.
How many other early gene disease association studies need to be checked?
Cathepsin C inhibitor chemical probe
As part of the Boehringer Ingelheim's efforts to foster innovation, they are share selected molecules with the scientific community all for free. The opnme portal gives access to a range of novel ligands. The latest addition is BI-9740
BI-9740 is a very potent and highly selective inhibitor of the enzymatic activity of Cathepsin C. It blocks human CatC in vitro with an IC50 of 1.8 nM and shows > 1500x selectivity versus the related proteases Cathepsin B, F, H, K, L and S. BI-9740 displays no activity against 34 unrelated proteases from different classes up to a concentration of 10 µM.
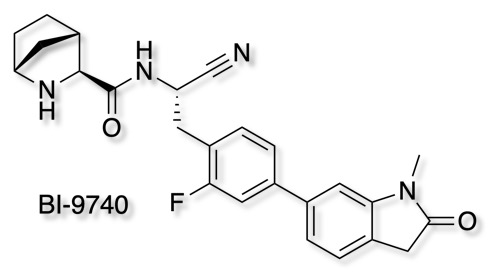
Chemical probes are absolutely essential for target validation and it is great to see so many high quality tools being made available.
Lysosome Targeting Chimeras (LYTACs)
A while back I added a page on PROteolysis Targeting Chimera (PROTAC) a technology using the ubiquitin proteasome system to induce degradation of the target protein DOI. However this technology is limited to cytosolic proteins.
A recent publication highlights a new technology "Lysosome Targeting Chimeras (LYTACs) for the Degradation of Secreted and Membrane Proteins" DOI that further extends the protein degradation options.
Targeted protein degradation is a powerful strategy to address the canonically undruggable proteome. However, current technologies are limited to targets with cytosolically-accessible and ligandable domains. Here, we designed and synthesized conjugates capable of binding both a cell surface lysosome targeting receptor and the extracellular domain of a target protein…. LYTACs represent a modular strategy for directing secreted and membrane proteins for degradation in the context of both basic research and therapy.
Website Update
I've spent some time over the last couple of weeks updating and adding new content to the Drug Discovery Resources section of the website.
In particular, s drug targets become more challenging medicinal chemists are looking at alternatives to small molecule competitive inhibitors, the section on covalent inhibitors have been expanded and a new page on PROTACs has been added. PROTACs are bifunctional molecules that bind to the target protein and an E3 ligase, the simultaneous PROTAC binding of two proteins brings the target protein in close enough proximity for polyubiquitination by the E2 enzyme associated to the E3 ligase, which flags the target protein for degradation through the proteasome. This offers a powerful alternative to competitive inhibition.
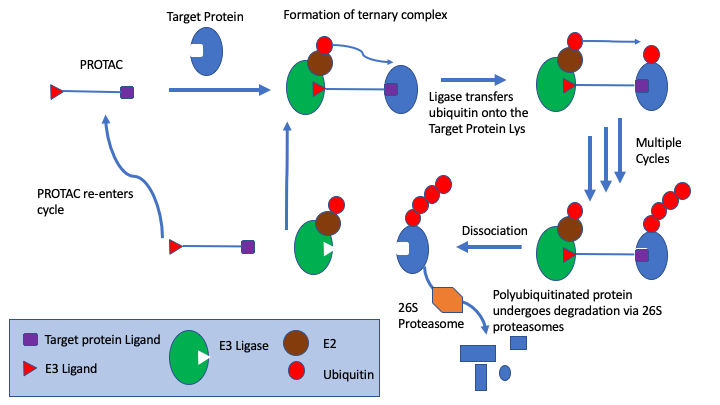
The Probes & Drugs portal has been added to the chemical probes page, this is a public resource joining together focused libraries of bioactive compounds (probes, drugs, specific inhibitor sets etc.) with commercially available screening libraries.
The page describing commercial fragment screening libraries has been updated to include a couple of new additions and flagging some that seem to be unavailable, if I've missed any feel free to let me know.
The section on hERG has been updated with links to new references and details of hERGcentral.
hERGCentral: A Large Database to Store, Retrieve, and Analyze Compound-Human Ether-à-go-go Related Gene Channel Interactions to Facilitate Cardiotoxicity Assessment in Drug Development. The hERGCentral database hergcentral.org is based on experimental data obtained from a primary screen by electrophysiology against more than 300,000 structurally diverse compounds screened at 1 and 10uM.
Unfortunately the database appears to be no longer available. Whilst the supplementary information for the original publication does not contain the structures of the tested compounds it does reference the PubChem substance ID. I used these identifiers to download the structures of the >300,000 records and combined them with the experimental data provided in the Excel tables in the supplementary information. The complete dataset can be downloaded from the hERG page.
Small molecules can potentially bind to a variety of bimolecular targets and whilst counter-screening against a wide variety of targets is feasible it can be rather expensive and probably only realistic for when a compound has been identified as of particular interest. For this reason there is considerable interest in building computational models to predict potential interactions the page on predicting bioactivities has been expanded.
The section on bioisosteres also have a few new examples.
Promises, promises, and precision medicine
A very interesting commentary on the impact (or lack of) genomics has had on human healthcare. J Clin Invest. 2019
The promises of precision medicine are to dramatically change patient care via individually tailored therapies and, as a result, to prevent disease, improve survival, and extend healthspan.
However, nearly two decades after the first predictions of dramatic success, we find no impact of the human genome project on the population’s life expectancy or any other public health measure, notwithstanding the vast resources that have been directed at genomics. Exaggerated expectations of how large an impact on disease would be found for genes have been paralleled by unrealistic timelines for success, yet the promotion of precision medicine continues unabated.
In light of the limitations of the precision medicine narrative, it is urgent that the biomedical research community reconsider its ongoing obsession with the human genome and reassess its research priorities including funding to more closely align with the health needs of our nation. We do not lack for pressing public health problems. We must counter the toll of obesity, inactivity, and diabetes; we need to address the mental health problems that lead to distress and violence; we cannot stand by while a terrible opiate epidemic ravages our country; we have to prepare conscientiously for the next influenza pandemic; we have a responsibility to prevent the ongoing contamination of our air, food, and water. Topics such as these have taken a back seat to the investment of the NIH and of many research universities in a human genome–driven research agenda that has done little to solve these problems, but has offered us promises and more promises.
The human genome project was undoubtedly a magnificent achievement, but has the investment in genomics delivered?
There is an extended discussion on In the Pipeline https://blogs.sciencemag.org/pipeline/archives/2019/01/31/precision-medicine-real-soon-now.
Open Targets Platform Updated
The latest update to the Open Targets Platform has been released (18.12).
Centre for Therapeutic Target Validation is a pre competitive public-private venture that aims to provide evidence on the biological validity of therapeutic targets and provide an initial assessment of the likely effectiveness of pharmacological intervention on these targets, using genome-scale experiments and analysis. The platform currently contains 28,931 targets, 3,049,882 associations for 10,053 diseases.
There is an open access portal to the platform here https://www.targetvalidation.org. All data is also available for download https://www.targetvalidation.org/downloads/data.
Another Chemical Probe
Boehringer Ingelheim have made a new addition to opnMe their portal for free chemical probes.
To foster innovation, we openly share selected molecules with the scientific community to unlock their full potential - all for free, no hidden costs.
The latest addition is a potent Chymase inhibitor, Chymase is a chymotrypsin-like serine protease that is stored in a latent form in the secretory granules of mast cells. Upon stimulation, it is released in its active form into the local tissue, contributing to the activation of TGF-ß, matrix metalloproteases and cytokines.
BI-1942 is a highly potent inhibitor of human chymase (IC50 = 0.4 nM) that can be used to test biological hypotheses involving this target in vitro. With BI-1829 we also offer a structurally close analog that is more than 1000 fold less active (IC50 = 850 nM) and can thus be used as negative control for in vitro studies.
More concerns about target validation
As a consultant I perhaps see more instances than most of the problems of reproducing literature studies, and I've highlighted several articles that have raised concerns. In particular, the concerns about antibody selectivity, the problems with irreproducible studies and the need for well characterised chemical probes. The excellent work by Elisabeth Bik looking at concerns with some of the images in the published literature, "The prevalence of inappropriate image duplication in biomedical research publications" mBio 7(3):e00809-16. DOI. her Twitter feed contains yet more examples from the current literature,
This latest correspondence in Nature highlights some of the issues, "Industry is more alarmed about reproducibility than academia" DOI.
This paragraph I find particularly troubling.
By contrast, academic scientists may be reluctant to devote extra time and effort to confirming research results in case they fail. That would put paid to publication in high-impact journals, damage career opportunities and curtail further funding. Evidence of questionable practices such as selective publishing and cherry-picking of data indicates that rigour is not always a high priority.
Drug Discovery Resources Updated
I've spent a little time updating the Drug Discovery Resources Section of the website. In particular:
- CYP interactions now includes details of published crystal structures and more information on known inhibitors
- I've added page on CYP1A2 from ChEMBL data and updated the CYP2D6 and CYP3A4 pages
- Updated the page on Aldehyde Oxidase
- Added new examples on the bioisosteres page
- Updated the Published Fragments section, including adding the overlay of all examples of Kinase fragment hits from the PDB.
- Added new examples to the Chemical Probes page
- Included more examples of halogen bonding to the Molecular Interactions page
Cyclophilin D as a drug target
Cyclophilin D (CYP D), is a member of a family of highly homologous peptidylprolyl cis-trans isomerases (PPIases) that interconvert the cis and trans isomers of peptide bonds with the amino acid proline. Proteins with prolyl isomerase activity include cyclophilins, FKBPs, and parvulin. Inhibitors of Cyclophilin D have been postulated as potential drugs for a variety of therapeutic targets including anti-viral activity DOI, neurodegenration DOI, Cancer DOI etc.
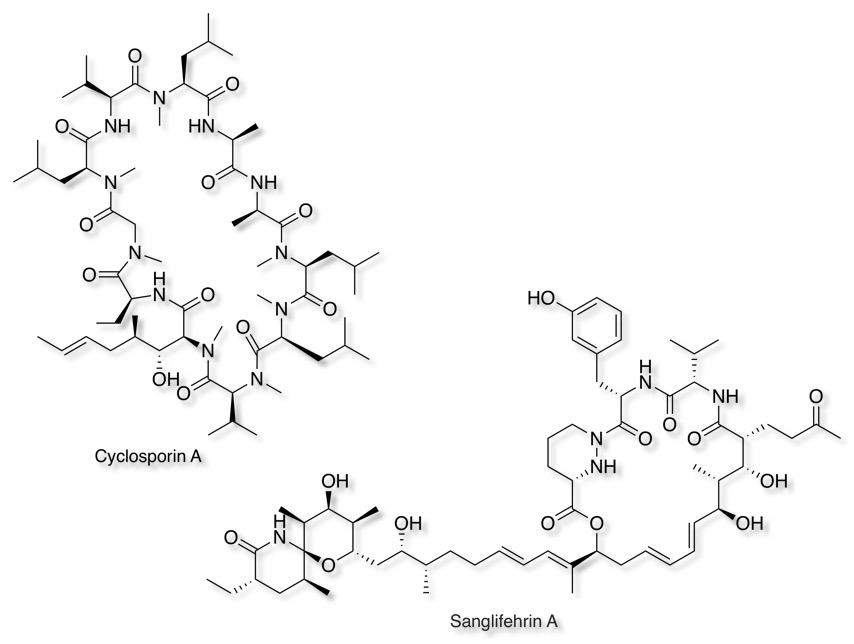
Until recently work in this area suffered from the lack of high quality, selective inhibitors, the best studied being the immunosuppressants Cyclosporin and Sanglifehrin A.
At the recent Macrocycles 2018 meeting Vicky Steadman described the identification and optimisation of potent and orally available selective Cyclophilin inhibitors, more details have just been published J Med Chem paper DOI.
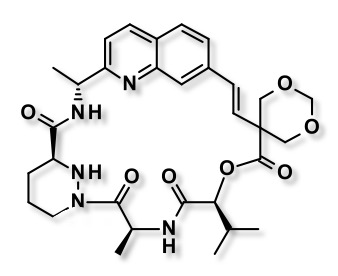
Let's hope with potent, cell penetrant and orally available tools in hand we can sort out the biology and bring forward a new class of therapeutic agents.
Open Targets Genetics
As mentioned on the Target Validation page, Open Targets is a public-private partnership that uses human genetics and genomics data for systematic drug target identification and prioritisation. The current focus is on oncology, immunology and neurodegeneration. An extension to this will be launched on 18 October 2018.
Open Targets Genetics is a new portal developed by Open Targets, an innovative partnership that brings together expertise from six complementary institutions to systematically identify and prioritise targets from which safe and effective medicines can be developed. The Portal offers three unique features to help you discover new associations between genes, variants, and traits, Gene2Variant analysis pipeline, Fine mapping/ credible set analysis, UK Biobank + GWAS Catalog integration.
More details https://genetics.opentargets.org/docs/open-targets-genetics-infographic.pdf
Open Targets Platform Update
Just announced. Open Targets Platform release - 18.08.
In this release there is now data on:
- 21,149 targets
- 10,101 diseases
- 6.5 million pieces of evidence
- 2.9 million associations between targets and diseases
The Open Targets Platform is a comprehensive and robust data integration for access to and visualisation of potential drug targets associated with disease. It brings together multiple data types and aims to assist users to identify and prioritise targets for further investigation. A drug target can be a protein, protein complex or RNA molecule and it’s displayed by its gene name from the Human Gene Nomenclature Committee, HGNC. We integrate all the evidence to the target using Ensembl stable IDs and describe relationships between diseases by mapping them to Experimental Factor Ontology (EFO) terms. The Platform supports workflows starting from a target or disease, and shows the available evidence for target – disease associations. Target and Disease profile pages showing specific information for both target (e.g baseline expression) and disease (e.g. Disease Classification) are also available
Open Molecules Platform
Boehringer Ingelheim has added it's well-characterised non-covalent ATP-competitive inhibitor of glycogen synthase kinase (GSK-3) Bi-5521 to its open molecule platform opnMe.com.
opnMe.com, the new open innovation portal of Boehringer Ingelheim, aims to accelerate research initiatives and enable new disease biology in areas of high unmet medical need by sharing well-characterized, best-in-class, pre-clinical tool compounds.
BI-5521 is a potent and selective ATP-competitive small molecule inhibitor of glycogen synthase kinase 3 (GSK-3), GSK-3β (IC50) 1.1 nM, with demonstrated in vivo activity. Rat pharmacokinetics are available, together with an inactive related compound.
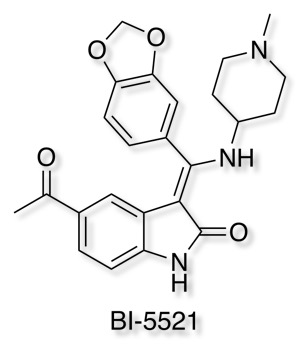
Another useful tool for Target Validation.
How reliable is the literature?
In the past I've mentioned some of the concerns about antibody selectivity, the problems with irreproducible studies and the need for well characterised chemical probes. Elisabeth Bik has been looking at concerns with some of the images in the published literature, The prevalence of inappropriate image duplication in biomedical research publications. mBio 7(3):e00809-16. DOI. her Twitter feed contains yet more examples from the current literature, well worth a browse.
So completing this set, I looked at 101 papers, all published in the same month in the same journal. Of these, 63 contained photographic images (the others had only line graphs and/or tables). Of those, 8 have potential duplications. That is 12.7%
With all the advances in AI and image recognition I'm slightly disappointed that there is not a programme that can do this automatically for Elisabeth.
Chemical Probes
Recently Boehringer Ingelheim have decided to provide access to a number of chemical probes.
Two new probes for BRD9 and BRD7/9 have been added.
I've added them to the Chemical Probes page.
Target Validation page updated
I've just updated the target validation page, highlighting some of the problems with the use of antibodies.
Neuroscience Chemical Probes
We recently heard that Pfizer is leaving the neuroscience therapeutic area, with a resulting loss of around 300 jobs. This is of course bad news for the scientists involved but I hope the work that was undertaken within Pfizer does not disappear. Chemical probes are critical tools in target identification and validation and arguably even more so in neuroscience. I hope that Pfizer consider releasing some of the well characterised molecules as freely accessible chemical probes, especially if they could also offer a similar but inactive molecule as a negative control. Many of the older tool compounds reported in the literature have been shown to have inadequate selectivity which compromises understanding the biology.
There are many important therapeutic targets within neuroscience but our biological understanding is currently inadequate to justify the investment in drug discovery, a selection of well characterised probes may provide the tools to support the necessary basic biological research.
opnMe Chemical probes from Boehringer Ingelheim
One of the key challenges to exploring interesting targets is having access to high quality molecular probes. A number of organisations have go together to support Chemical Probes Portal which provides information and independent reviews of chemical probes.
The Chemical Probes Portal is designed to change the way scientists find and use small-molecule reagents called chemical probes in biomedical research and drug discovery. The Portal is backed by reviews and commentary from recognised chemical probe experts. Our knowledge-dissemination model, focused on providing accessible expert advice, promises to increase research reproducibility, maximise investment outcomes and accelerate the discovery science that informs the next generation of therapeutic
Recently Boehringer Ingelheim have decided to provide access to a number of chemical probes.
To foster innovation, Boehringer Ingelheim (BI) is openly sharing selected molecules with the scientific community to unlock their full potential. There are two types of Boehringer Ingelheim molecules that you can access on this portal: some for ordering, some for collaboration.
These molecules cover a range of interesting molecular targets.
| Target | ID |
|---|---|
| Aurora B inhibitor | BI 831266 |
| Autotaxin (ATX) inhibitor | BI-2545 |
| BCL6 degrader | BI-3802 |
| BCL6 inhibitor | BI-3812 |
| CCR1 antagonist | BI-9667 |
| CCR10 antagonist | BI-6901 |
| CDK8 inhibitor | BI-1347 |
| FAS inhibitor | BI 99179 |
| FLAP antagonist | BI 665915 |
| Glucocorticoid Receptor (GR) Agonist | BI 653048 |
| Hep. C virus (HCV) NS5B polymerase inhibitor | BI 207127 (Deleobuvir) |
| Hepatitis C virus (HCV) NS3 protease inhibitor | BI-1230 |
| Hepatitis C virus (HCV) NS3 protease inhibitor | BI-1388 |
| LFA-1 (lymphocyte function-associated antigen-1) antagonist | BI-1950 |
| NHE1 inhibitor | BI-9627 |
| PLK1 inhibitor | BI-2536 |
| sEH inhibitor | BI-1935 |
| SYK inhibitor | BI 1002494 |
Looking at the selective Aurora B kinase inhibitor BI 831266, it is clear that BI is making available high quality molecules, they provide the structure, in vitro activity, together with both in vitro and in vivo DMPK data in multiple species. They also suggest a related compound as a negative control in which the N-Me serves to block the critical hinge binding.
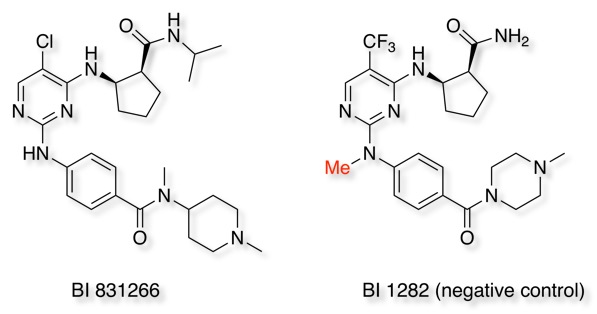
There is also a co-crystal structure and some counter-screening data, together with key references from the literature. Any data generated can be published without approval from BI.
This looks to be a very exciting initiative and it will be interesting to see if other companies follow suit.
They have also created a search engine BI Miner to search multiple data sources simultaneously (PubMed Central, Medline, Patents, Drug labels, Expression Data, NIH Grants, Clinical Trials), this open access.
Trim-Away
A new way to deplete endogenous proteins, Trim-Away a technique to degrade endogenous proteins acutely in mammalian cells without prior modification of the genome or mRNA. Trim-Away harnesses the cellular protein degradation machinery to remove unmodified native proteins within minutes of application.
We reasoned that the antibody receptor and ubiquitin ligase TRIM21 could be used as a tool to drive the degradation of endogenous proteins by using a 3-step strategy: first, the introduction of exogenous TRIM21; second, the introduction of an antibody against the protein of interest; and third, TRIM21-mediated ubiquitination followed by degradation of the antibody-bound protein of interest.
There is more information on the MRC website
I've added it to the Target Validation page.
Updated Target Validation
I've updated the Target Identification/Validation page in the Drug Discovery Resources. In particular I've added a section on the failure to reproduce literature experiments.
Remember this is an absolutely critical step, almost everything else can be fixed.
Target validation and antibodies
A couple of years ago I mentioned an article reviewing antibody selectivity
In 2011, an evaluation of 246 antibodies used in epigenetic studies found that one-quarter failed tests for specificity, meaning that they often bound to more than one target. Four antibodies were perfectly specific — but to the wrong target.
The issue of antibody selectivity has again been flagged as a concern in oestrogen receptor beta research DOI. This is a major target for breast cancer research and there are multiple ongoing clinical trials https://clinicaltrials.gov/ct2/results?term=ERβ.
We here perform a rigorous validation of 13 anti-ERβ antibodies, using well-characterized controls and a panel of validation methods. We conclude that only one antibody, the rarely used monoclonal PPZ0506, specifically targets ERβ in immunohistochemistry.
Applying this antibody for protein expression profiling in 44 normal and 21 malignant human tissues, we detect ERβ protein in testis, ovary, lymphoid cells, granulosa cell tumours, and a subset of malignant melanoma and thyroid cancers. We do not find evidence of expression in normal or cancerous human breast.
Perhaps more worryingly the authors comment.
While our study focuses on ERβ, we do not think that antibodies towards ERβ are significantly poorer than those targeting other proteins, and it is not unlikely that this problem generates similar obstacles in many other fields.
As I wrote on the Target Validation page
Remember this is an absolutely critical step, almost everything else can be fixed.
Target Validation or not
The reproducibility of some target identification/validation studies has been questioned on several occasions and I've flagged up some of the concerns in the Target Validation section of the Drug Discovery Resources. A recent study, reported in Science 28 August 2015: Vol. 349 no. 6251 DOI looking at psychological science, attempted to replicate published work suggests that 39% of effects replicated the original result. Also Amgen, tried to replicate 53 'landmark' cancer studies and failed to replicate the original studies in all but six occasions, Nature 483, 531–533 (29 March 2012) DOI.
A while back a project was initiated to look at reproducibility in cancer, Science Forum: An open investigation of the reproducibility of cancer biology research DOI.
It is widely believed that research that builds upon previously published findings has reproduced the original work. However, it is rare for researchers to perform or publish direct replications of existing results. The Reproducibility Project: Cancer Biology is an open investigation of reproducibility in preclinical cancer biology research. We have identified 50 high impact cancer biology articles published in the period 2010-2012, and plan to replicate a subset of experimental results from each article. A Registered Report detailing the proposed experimental designs and protocols for each subset of experiments will be peer reviewed and published prior to data collection. The results of these experiments will then be published in a Replication Study. The resulting open methodology and dataset will provide evidence about the reproducibility of high-impact results, and an opportunity to identify predictors of reproducibility.
Well some of the early results are in and they make for pretty sobering if not unexpected reading, of the first 7 papers examined, 2 appear to reproduce the original finding to some extent, three show significant differences from the original studies. The results are published in eLife here and there is an editorial here DOI, and they make an important point.
if all the original studies were reproducible, not all of them would be found to be reproducible, just based on chance. The experiments in the Reproducibility Project are typically powered to have an 80% probability of reproducing something that is true.
The key question is of course, is the failure to reproduce these results due to methodological differences not apparent from the described experimental or whether the fundamental result is invalid. At the moment if you are planning to invest in a drug discovery project based on a single publication then Caveat emptor.
47 de-prioritised pharma compounds opens to researchers
A collection of 47 deprioritised pharmaceutical compounds and up to £5 million is being made available to academic researchers through the latest round of the MRC-Industry Asset Sharing Initiative. The collaboration, between the MRC and six global drug companies, is the largest of its kind in the world.
The list of compounds is available here together with brief description of the molecular target and pharmacology, Safety, Tolerability and toxicity information, and any clinical studies that might have been undertaken.
Target Valaidation site update
TargetValidation.org has been updated.
This release brings new web displays and plenty of extra data to assist you in drug discovery and validation:
- 30,591 targets
- 9,425 diseases
- 4.8 million evidence
- 2.4 million target-disease associations
There are also new Web Widgets for both 'RNA baseline expression' and 'Protein Structure' of a target. In the latter, you can now rotate the protein structure, change its colour, zoom in and out, and highlight any amino acid residue:
More information is available on the blog
Chemical Probes Portal Updated
The Chemical Probes Portal has been updated, the new site includes a lot more data about the existing probes, reviewer ratings and their comments.
A chemical probe is simply a reagent—a selective small-molecule modulator of a protein’s function—that allows the user to ask mechanistic and phenotypic questions about its molecular target in cell-based and/or animal studies. These are tools not drugs, they allow scientists to investigate the relationship between a molecular target and the broader biological consequences of modulating that target in cells or organisms. In general the focus is on specificity for the target rather than pharmacokinetics.