Chemical Probes
Whilst there may be known small molecules that exert the desired effect but perhaps with a sub-optimal profile. It is absolutely critical to use good quality probes to avoid generating misleading results. Whilst chemical probes are probably mainly thought of to provide evidence of on-target function and translation into a therapeutic effect they can also be important for de-risking potential on-target toxicity,
It is important to note that chemical probes are not potential drug candidates, the critical need is for selectivity, both for the parent compound and potential metabolites. Oral bioavailability is not essential and the availability of inactive analogues (perhaps enantiomers) is highly desirable. A knowledge of the probe pharmacokinetics is however useful since it can give an insight into the PK/PD relationship. Unfortunately not all chemical probes are well characterised, in addition over time it may become apparent that a molecule also exhibits additional activities. For this reason the http://www.chemicalprobes.org has been developed. The database currently contains nearly 200 different probes rated by SAB members on a scale 1-4 stars.
4 stars = Recommended as a probe for this target
3 stars = Best available probe for this target, or a high quality probe that is a useful orthogonal tool
2 stars = Insufficient validation data to recommend
1 star = Not recommended as a probe for this target
Some examples of chemical probes are shown below, together with the average rating and the number of reviews. The full listing is available here
| Probe Name | Protein target | Target class | Subclass | Number of SAB Reviews | Avg Rating (in cells) |
|---|---|---|---|---|---|
| Taxol | Beta-Tubulin | Structural protein | Cytoskeleton | 1 | 4.0000 |
| PFI-1 | BRD2, BRD3, BRD4, BRDT | Epigenetics | Bromodomain | 3 | 4.0000 |
| FM-381 | JAK3 | Protein kinase | Tyrosine kinase | 3 | 4.0000 |
| UNC1215 | L3MBTL3 | Epigenetics | Malignant brain tumor Reader | 3 | 4.0000 |
| MI-77301 | MDM2 | Other post-translational modification | E3 ubiquitin ligase | 3 | 4.0000 |
It is also important to note that a resource like chemicalprobes.org is hand curated by experts and relies on contributions from many labs.
This resource is a community driven wiki-like site that recommends appropriate chemical probes for biological targets, provides guidance on their use, and documents their limitations. We also provide advice on the use of controls, both chemically distinct probes for the same target, and negative control compounds, where available.
It may also be useful to be able to compare CNS penetrant and non-CNS penetrant probes for some indications. It may be possible to identify potential liabilities at this point or key off-targets issues. Put a screening assay in place together with functional screen.
In contrast Probe Miner is a resource for the evaluation of chemical probes built on large-scale, publicly available, medicinal chemistry data that allows objective analysis of potential probes. DOI. This resource uses data on nearly two million molecules against 2,220 human targets. However this served to underline the paucity of publicly available high quality data.
From the 1.8 million total compounds (TC) available in public databases, we find that only 355,305 human active compounds (HAC) have some acceptable level of biochemical activity (<10 μM; see STAR Methods) reported against a human protein. Of these, 189,736 (10.5% TC, 53% HAC) have measured biochemical activity or binding potency of 100 nM or better. However, when considering selectivity, we find that only 93,930 compounds have reported binding or activity measurements against two or more targets. Of these, only 48,086 (2.7% TC, 14% HAC) satisfy both our minimal potency and selectivity criteria
To create a metric that allows objective, data-driven ranking of all compounds tested for a particular protein target, they developed a set of six scores using previously described fitness factors DOI. The key scoring properties are Potency Score, Selectivity Score, Cell Score, Structure-Activity Relationship (SAR) Score, Inactive Analog Score, and PAINS Score (see table below). By using this scoring scheme they can highlight those molecules that are likely to be suitable as probes.
| Property | Description |
|---|---|
| Target Selectivity | It shows whether a compound inhibiting this protein is screened against at least one other target and has at least 10-fold selectivity against any other target |
| Target Potency | It shows whether a compound inhibits this target with at least 100 nM potency |
| Cell Potency | It shows whether a compound binding to the target of interest is active in a cell line with at least 10 μM potency |
| Minimum Standard | It is an aggregate of the three previous scores (which themselves are independent from each other), indicating whether there are compounds inhibiting this target with minimum standards of target potency (pActivity ≥7), selectivity (at least one tested off-target and 10-fold selectivity against off-targets) and cell potency (activity below 10 μM in at least one cell line) simultaneously. It is a key icon showing whether a compound fulfilling these minimum-quality requirements is found in publicly available databases |
| SAR | It indicates that there is at least one compound binding to this target that has SAR as defined by the SAR Score (see above) |
| Inactive Analog | It indicates that there is at least one inactive analog of the compound as defined by the Inactive Analog Score (see above) |
| PAINS | It shows that there is at least one compound inhibiting this target that has no PAINS alerts as defined in the PAINS Score |
The Probes & Drugs portal is a public resource joining together focused libraries of bioactive compounds (probes, drugs, specific inhibitor sets etc.) with commercially available screening libraries. The purpose of the portal is to reflect the current state of bioactive compound space and to enable its exploration from different points of view DOI.
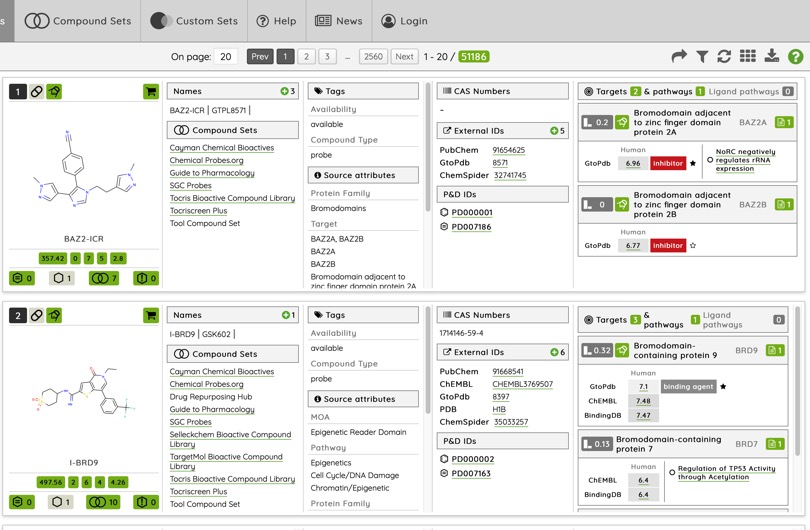
The portal provides detailed information on a wide variety of molecules as shown below.

Recently Boehringer Ingelheim have decided to provide access to a number of chemical probes.
To foster innovation, Boehringer Ingelheim (BI) is openly sharing selected molecules with the scientific community to unlock their full potential. There are two types of Boehringer Ingelheim molecules that you can access on this portal: some for ordering, some for collaboration.
These molecules cover a range of interesting molecular targets and more molecules are added regularly.
| Target | ID | structure |
|---|---|---|
| Aurora B inhibitor | BI 831266 | 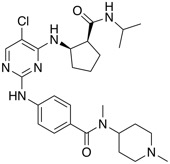 |
| Autotaxin (ATX) inhibitor | BI-2545 | 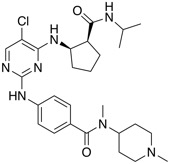 |
| BCL6 degrader | BI-3802 | 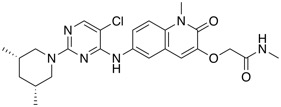 |
| BCL6 inhibitor | BI-3812 | 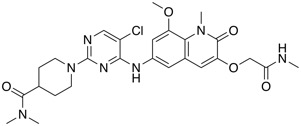 |
| BRD9 inhibitor | BI-9564 | 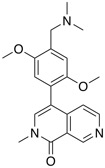 |
| BRD7/9 inhibitor | BI-7373 | 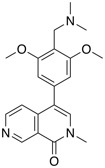 |
| CCR1 antagonist | BI-9667 | 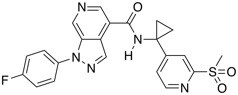 |
| CCR10 antagonist | BI-6901 | 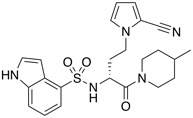 |
| CDK8 inhibitor | BI-1347 | 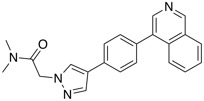 |
| FAS inhibitor | BI 99179 | 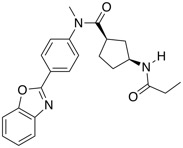 |
| FLAP antagonist | BI 665915 | 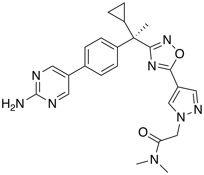 |
| Glucocorticoid Receptor (GR) Agonist | BI 653048 | 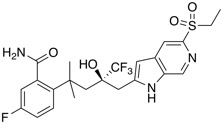 |
| Hep. C virus (HCV) NS5B polymerase inhibitor | BI 207127 (Deleobuvir) | 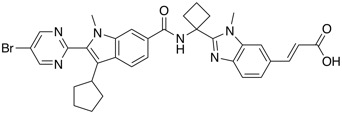 |
| Hepatitis C virus (HCV) NS3 protease inhibitor | BI-1230 | 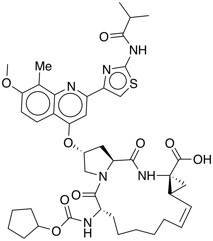 |
| Hepatitis C virus (HCV) NS3 protease inhibitor | BI-1388 | 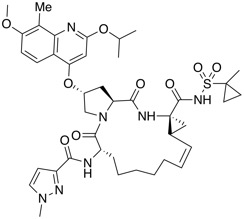 |
| LFA-1 (lymphocyte function-associated antigen-1) antagonist | BI-1950 | 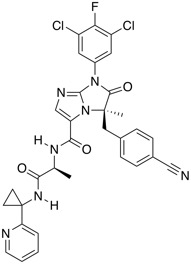 |
| NHE1 inhibitor | BI-9627 | 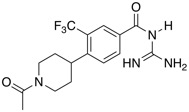 |
| PLK1 inhibitor | BI-2536 | 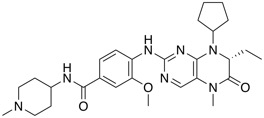 |
| sEH inhibitor | BI-1935 | 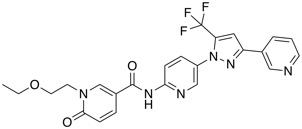 |
| SYK inhibitor | BI 1002494 | 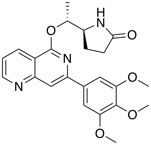 |
| Chymase inhibitor | BI 1942 | 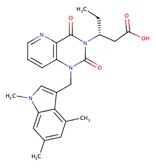 |
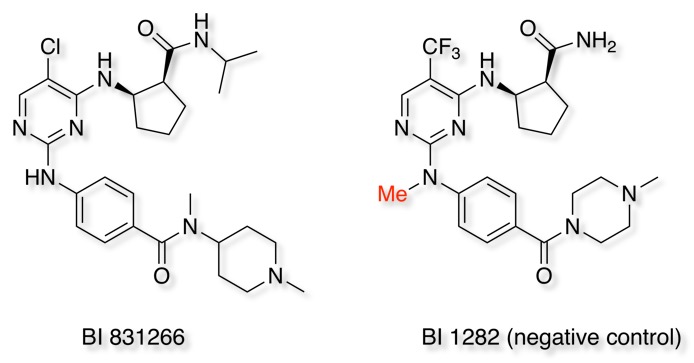
Looking at the selective Aurora B kinase inhibitor BI 831266, it is clear that BI is making available high quality molecules, they provide the structure, in vitro activity, together with both in vitro and in vivo DMPK data in multiple species. They also suggest a related compound as a negative control in which the N-Me serves to block the critical hinge binding.
Worth reading
The promise and peril of chemical probes DOI.
The Impact of Chemical Probes in Drug Discovery: A Pharmaceutical Industry Perspective DOI.
Will the chemical probes please stand up? DOI.
Last updated 22 December 2022