EU restrictions on Fluorochemicals
In drug discovery the introduction of fluorine into potential drug candidates is an essential part of the medicinal chemists toolbox. Blocking a metabolic hotspot by replacing a Hydrogen by Fluorine or deactivating and aromatic ring by adding a trifluoromethyl are well established strategies for reducing metabolism, increasing half-life and reducing drug load. There are more details on the metabolism page and influence [pi-stacking interactionshttps://www.cambridgemedchemconsulting.com/resources/molecular_interactions.html).
I have a database of all drugs that have been reported to be in phase 3 trials or later (not all will have made it to market) and it is interesting to see how many contain a fluorine atom of some kind. As expected Aryl F and CF3 are the most common
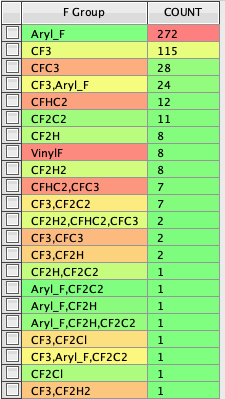
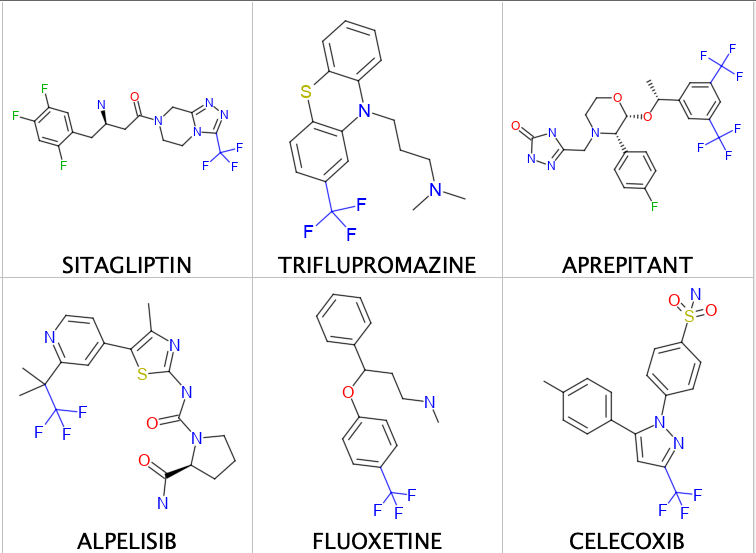
A few examples are shown below.
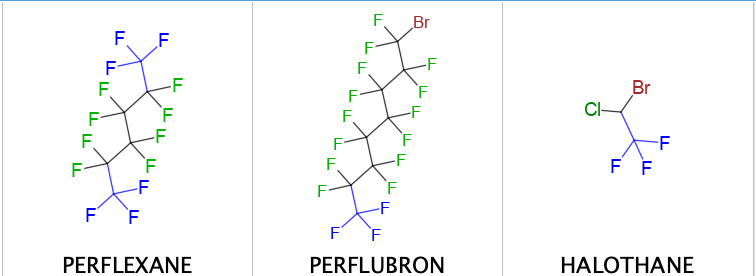
There are also other classes of molecules like perflexane which is used as an imaging contrast agent in echocardiogram. Halothane used clinically as an inhalational anesthetic (on the WHO Model List of Essential Medicines) and other inhaled anesthetics.
So at first sight the EFMC statement was of some concern.
EFMC STATEMENT ON EU PROPOSAL TO BAN PFAS The EU, through the European Chemicals Agency (ECHA), has launched a proposal that aims to extensively restrict the manufacture, supply, and use of all per- and polyfluoroalkyl substances (Ref 1). PFAS are a large class of synthetic chemicals that are used across a broad range of activities in different scientific areas, not restricted to chemistry. However, several PFAS are environmental pollutants and some of them have detrimental effects on human health. Their extensive use throughout the society, combined with the low reactivity displayed by many fluoroalkyl chemicals, magnifies the potential for accumulation in the environment and contamination of food and water supplies. According to this proposal, which is in public consultation until September 25th (ref 2), PFAS encompasses "any substance that contains at least one fully fluorinated methyl (CF3) or methylene (CF2) without any H, Cl, Br, or I attached to it"
However, looking at the generic scope in more detail it appears that the scope might not be as all encompassing. Looking at the description here https://echa.europa.eu/registry-of-restriction-intentions/-/dislist/details/0b0236e18663449b. The generic scope is described as shown below and I've highlighted a critical phrase.
Per- and polyfluoroalkyl substances (PFASs) defined as: Any substance that contains at least one fully fluorinated methyl (CF3-) or methylene (-CF2-) carbon atom (without any H/Cl/Br/I attached to it).
A substance that only contains the following structural elements is excluded from the scope of the proposed restriction:
CF3-X or X-CF2-X’,
where X = -OR or -NRR’ and X’ = methyl (-CH3), methylene (-CH2-), an aromatic group, a carbonyl group (-C(O)-), -OR’’, -SR’’ or –NR’’R’’’,
and where R/R’/R’’/R’’’ is a hydrogen (-H), methyl (-CH3), methylene (-CH2-), an aromatic group or a carbonyl group (-C(O)-).
My interpretation (caveat I'm not a lawyer) is that the vast majority of drugs, reagents and solvents like trifluoro acetic acid would be excluded. However, inhaled anesthetics and the contrast imaging agents would be included. So the pharma industry needs to be included in the dialog but drugs themselves might have limited impact.
I've not covered polymers, and PTFE is is used in much laboratory equipment from stirrer bars, stopcocks, O-rings, seals etc. and who hasn't used PTFE tape on ground glass joints. There may be alternatives that I'm not aware of, but any substance that has similar properties of inertness, durability etc. may cause the same concerns as PFAs.
The European Chemicals Agency invites interested parties to send in scientific and technical information on the manufacture, placing on the market and use of per- and polyfluoroalkyl substances (PFAS) by 25 September 2023 https://echa.europa.eu/-/echa-seeks-input-on-proposed-pfas-restriction.
Predicting sites of metabolism
I updated the page on predicting metabolism
https://www.cambridgemedchemconsulting.com/resources/ADME/predicting_metabolism.html.
Updated Drug Discovery Resources
Updated the page on metabolism https://www.cambridgemedchemconsulting.com/resources/ADME/metabolism.html.
And the page on covalent ligands https://www.cambridgemedchemconsulting.com/resources/lead_identification/covalent.html.
Updated the page on reducing metabolism
Updated the page on reducing metabolism, included example of using deuterium to block metabolism in a compound in the clinic.
Predicting sites of metabolism
I've updated the predicting sites of metabolism page
https://www.cambridgemedchemconsulting.com/resources/ADME/predicting_metabolism.html
Predicting sites of metabolism
I've added GLORYx to the predicting metabolism page.
GLORYx predicts phase I and phase II metabolites for the chemical compound(s) provided by the user. The method is based on the FAME site of metabolism (SoM) prediction combined with sets of reaction rules encoding both phase I and phase II metabolic reactions.
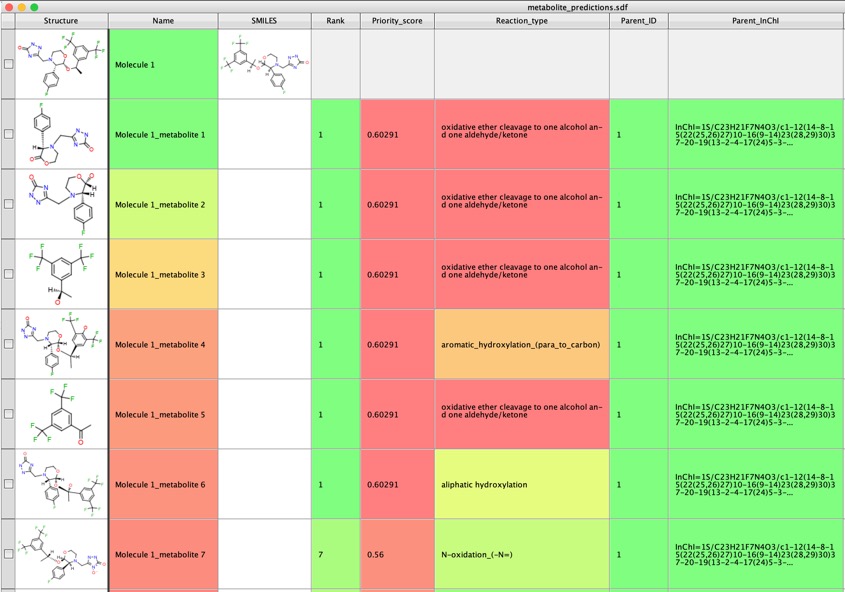
I tried a range of other molecules and GLORYx was really very impressive in identifying potential metabolites.
Updated Drug Discovery Resources
I've done some updates to the Drug Discovery Resources.
The Following Pages have been Updated
Macrocycles
Predicting Metabolism
Covalent Inhibitors
PROteolysis TArgeting Chimeras (PROTACs), Lysosome Targeting Chimeras (LYTACs), and ENDosome TArgeting Chimeras (ENDTACs)
Predicting Sites of Metabolism page updated
I've updated the Predicting sites of metabolism page.
Predicting sites of metabolism
I have updated the drug discovery resources on predicting sites of metabolism, I've added several new tools and web-based resources.
Aldehyde Oxidase page updated
I’ve updated the page on Aldehyde Oxidase, an enzyme in metabolism of a wide variety of nitrogen heterocycles.
I’ve also included A recent publication DOI that suggests a simple test for the early identification of heteroaromatic drug candidates that have a high probability of metabolism by AO. Bis(((difluoromethyl)sulfinyl)oxy)zinc (DFMS) was used as a source of the CF2H racial, simple LCMS was then used to identify a characteristic M+50 peak. It is also possible to scale up and isolate these metabolically blocked compounds and retest them for improved qualities.
Updates
I’ve added a sections on fragment-based screening, solubility and updated the section on metabolism.