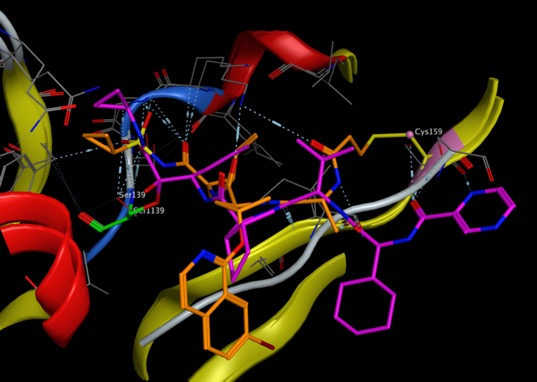
Covalent Inhibitors
Covalent Inhibitorrs are an increasingly investigated class of drugs. The attraction is the irreversible nature of the binding and subsequent prolonged target engagement. An invaluable resource for this area of research is the Covalent Inhibitor Database that has recently been updated to version 2.0.
This updated version includes 8303 inhibitors and 368 targets, supplemented by 3445 newly added cocrystal structures, providing detailed analyses of non-covalent interactions.
The article describing the database is here DOI.
BMCS Hot Topics Meetings
The next meeting is Hot Topics: Covalent Drug Discovery 2024, this online event is on Thursday 16th May, 2024 (afternoon).
To register for the meeting, click here To download the first announcement poster, click here
Binding Sites are 3D
I've always found it interesting that whilst everyone recognises that protein binding sites are three dimensional (and chiral) there is a reluctance to have chiral centres in screening hits. This is despite examples were chiral centres aid affinity, selectivity and solubility. I suspect one of the concerns is the ease of follow up for any hits.
I've been working with Liverpool Chirochem to design a 3D rich, homochiral fragment screening library. The real beauty of this library is that the fragments can be easily expanded using validated chemistry in their parallel synthesis lab.
Once you have built a supply of these homochiral building blocks they can of course be put to many different additional uses, covalent fragments, DEL building blocks, and as building blocks for large virtual libraries. All of which can be supported by their parallel synthesis lab.
Updated Drug Discovery Resources
I have updated several pages in the Drug Discovery Resources.
Ester and Amide Bioisosteres https://www.cambridgemedchemconsulting.com/resources/bioisoteres/ester_bioisosteres.html
Cysteine Protease inhibitors https://www.cambridgemedchemconsulting.com/resources/hitidentification/focus/cysteineprotease_inhibitors.html Covalent Inhibitors https://www.cambridgemedchemconsulting.com/resources/lead_identification/covalent.html.
A database of covalent binders
A recent publication describes CovBinderDB DOI is a fantastic resource mined from the PDB that contains 7375 covalent modifications in which 2189 unique covalent binders target nine types of amino acid residues (Cys, Lys, Ser, Asp, Glu, His, Met, Thr, and Tyr) from 3555 complex structures of 1170 unique protein chains. The database can be accessed here https://yzhang.hpc.nyu.edu/CovBinderInPDB/.
I've added it to the Covalent inhibitors page of the Drug Discovery Resources.
Updated Drug Discovery Resources
Updated the page on metabolism https://www.cambridgemedchemconsulting.com/resources/ADME/metabolism.html.
And the page on covalent ligands https://www.cambridgemedchemconsulting.com/resources/lead_identification/covalent.html.
Computational Prediction of covalent Inhibitors
Covalent Inhibitors are an increasingly important class of therapeutic agents.
A computational pipeline has been described by the London lab to predict suggest covalent analogs of non-covalent ligands DOI.
Designing covalent inhibitors is increasingly important, although it remains challenging. Here, we present covalentizer, a computational pipeline for identifying irreversible inhibitors based on structures of targets with non-covalent binders. Through covalent docking of tailored focused libraries, we identify candidates that can bind covalently to a nearby cysteine while preserving the interactions of the original molecule. We found ∼11,000 cysteines proximal to a ligand across 8,386 complexes in the PDB. Of these, the protocol identified 1,553 structures with covalent predictions. In a prospective evaluation, five out of nine predicted covalent kinase inhibitors showed half-maximal inhibitory concentration (IC50) values between 155 nM and 4.5 μM. Application against an existing SARS-CoV Mpro reversible inhibitor led to an acrylamide inhibitor series with low micromolar IC50 values against SARS-CoV-2 Mpro. The docking was validated by 12 co-crystal structures. Together these examples hint at the vast number of covalent inhibitors accessible through our protocol.
RDKit was used for 2D molecular handling, conformation generation and RMSD calculation. RDKit: Open-source cheminformatics; version 2018.09.3; RDKit.org. Marvin was used in the process of preparing the molecules for docking, Marvin 17.21.0, ChemAxon (https://www.chemaxon.com). OpenBabel (http:// openbabel.org/wiki/Main_Page) was used to switch between molecular file formats. DOCKovalent (London et al., 2014) was used for virtual covalent docking. The Covalentizer code is available at https://github.com/LondonLab/Covalentizer.
Covalent Modification
I've written a page in the Drug Discovery Resources on covalent inhibitors.
This publication suggests this area is going to become more important "An activity-guided map of electrophile-cysteine interactions in primary human immune cells" https://www.biorxiv.org/content/10.1101/808113v1.
Here, we present a global map of cysteines in primary human T cells that are susceptible to covalent modification by electrophilic small molecules. More than 3000 covalently liganded cysteines were found on functionally and structurally diverse proteins, including many that play fundamental roles in immunology.
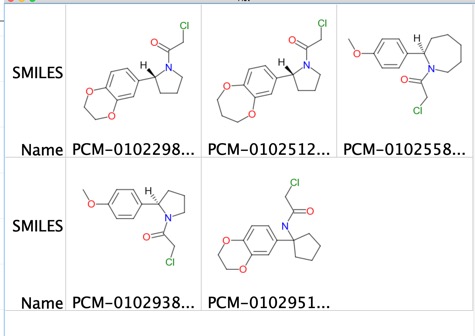
Covalent Inhibtors page updated
I've updated the covalent inhibitors page to include details of two covalent fragment libraries that have been used in fragment screening.
Updated Drug Discovery Resources
I've done some updates to the Drug Discovery Resources.
The Following Pages have been Updated
Macrocycles
Predicting Metabolism
Covalent Inhibitors
PROteolysis TArgeting Chimeras (PROTACs), Lysosome Targeting Chimeras (LYTACs), and ENDosome TArgeting Chimeras (ENDTACs)
Drug Discovery Resources Updated
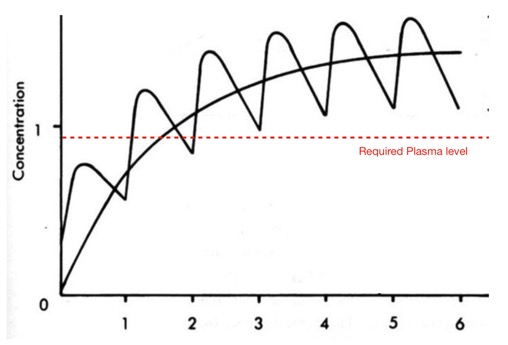
I've made a few additions and updated to the Drug Discovery Resources pages. In particular I've updated the covalent inhibitors page and added additional examples to the molecular interactions page. I've also started updating the ADME section and added a page on half-life and how it might be modulated.
Covalent Inhibitors
I've just added a page on irreversible covalent inhibitors, this is just the first iteration I plan to add more when I have some spare time.