Selecting Compounds from a Virtual Screening Run
Whilst high-throughput screening (HTS) has been the starting point for many successful drug discovery programs the cost of screening, the accessibility of a large diverse sample collection, or throughput of the primary assay may preclude HTS as a starting point and identification of a smaller selection of compounds with a higher probability of being a hit may be desired. Directed or Virtual screening is a computational technique used in drug discovery research designed to identify potential hits for evaluation in primary assays. It involves the rapid in silico assessment of large libraries of chemical structures in order to identify those structures that most likely to be active against a drug target. The key question is then how many molecules do you select from your virtual screen?
The results of a virtual screening run are effectively a rank ordering of the virtual screening deck ordered by whatever scoring function(s) that have been used. The task then becomes selection of molecules for experimental determination of activity.
I posed this question on the website and the results are shown below. Whilst this obviously a limited snapshot it is interesting that there is a wide variety of responses.
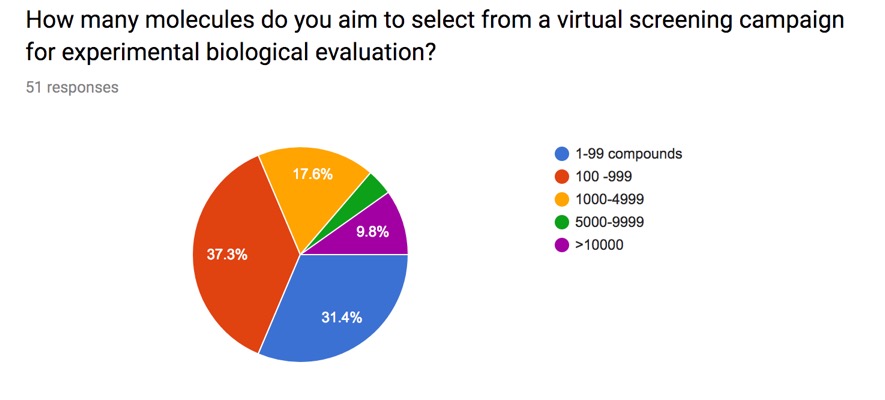
Some people also emailed me with further information. For companies with large internal physical screening collections, and the ability to cherry pick samples, it effectively costs the same to fill a high density plate (>1000 compounds) as it does to select a handful of compounds. On the other hand if the scientist has to purchase compounds then the logistics and cost become a significant obstacle. It would have been interesting to compare different virtual screening techniques, academic versus biotech versus large pharma etc. but I doubt I'd get as many answers from a multi-page questionnaire.
There is an interesting publication "Predictiveness curves in virtual screening" by Charly Empereur-mot et al DOI in which they look compare several docking methods and use the predictiveness curve as a quantification of the predictive performance of virtual screening methods on a fraction of a given molecular dataset. They use the Directory of Useful Decoys datasets (DUD) for comparison and were kind enough to provide me with the results, I've just used the data generated using Autodock Vina.
DUD consists of a total of 2,950 active compounds against a total of 40 targets. For each active, 36 "decoys" with similar physical properties (e.g. molecular weight, calculated LogP) but dissimilar topology
Compared to the typical results of high-throughput screening where the hit rate is usually <1%, as the table below shows DUD contains an unusually high concentration of actives (2-5%), but the results of the virtual screening are certainly very informative.
| Target | No. of actives | No. of compounds | Prevalence |
|---|---|---|---|
| ACE | 49 | 1846 | 0.0265 |
| ACHE | 107 | 3999 | 0.0268 |
| ADA | 39 | 966 | 0.0404 |
| ALR2 | 26 | 1021 | 0.0255 |
| AMPC | 21 | 807 | 0.0260 |
| AR | 79 | 2933 | 0.0269 |
| CDK2 | 72 | 2146 | 0.0336 |
| COMT | 11 | 479 | 0.0230 |
| COX-1 | 25 | 936 | 0.0267 |
| COX-2 | 426 | 13715 | 0.0311 |
| DHFR | 410 | 8777 | 0.0467 |
| EGFR | 475 | 16471 | 0.0288 |
| ER ago | 67 | 2637 | 0.0254 |
| ER antago | 39 | 1487 | 0.0262 |
| FGFR1 | 120 | 4670 | 0.0257 |
| FXA | 146 | 5891 | 0.0248 |
| GART | 40 | 919 | 0.0435 |
| GPB | 52 | 2192 | 0.0237 |
| GR | 78 | 3025 | 0.0258 |
| HIVPR | 62 | 2100 | 0.0295 |
| HIVRT | 43 | 1562 | 0.0275 |
| HMGR | 35 | 1515 | 0.0231 |
| HSP90 | 37 | 1016 | 0.0364 |
| INHA | 86 | 3352 | 0.0257 |
| MR | 15 | 651 | 0.0230 |
| NA | 49 | 1923 | 0.0255 |
| P38 | 454 | 9595 | 0.0473 |
| PARP | 35 | 1386 | 0.0253 |
| PDE5 | 88 | 2066 | 0.0426 |
| PNP | 50 | 1086 | 0.0460 |
| PPAR | 85 | 3212 | 0.0265 |
| PR | 27 | 1068 | 0.0253 |
| RXR | 20 | 770 | 0.0260 |
| SAHH | 33 | 1379 | 0.0239 |
| SRC | 159 | 6478 | 0.0245 |
| THR | 72 | 2528 | 0.0285 |
| TK | 22 | 913 | 0.0241 |
| TRP | 49 | 1713 | 0.0286 |
| VEGFR2 | 88 | 2994 | 0.0294 |
| Minimum | 11 | 479 | 0.0230 |
| Maximum | 475 | 16471 | 0.0473 |
| Mean | 97 | 3134 | 0.0294 |
| Median | 50 | 1923 | 0.0265 |
Table 1 shows a summary of the partial metrics at 2% and 5% of the ordered dataset for virtual screens performed using Autodock Vina, partial total gain (pTG), partial area under the curve (pAUC), Enrichment factors (EF)
| Table 1 | Autodock Vina – Top 2% dataset | Autodock Vina – Top 5% dataset | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target | pTG 2% | pAUC 2% | EF 2% | Actives 2% | Cpds 2% | pTG 5% | pAUC 5% | EF 5% | Actives 5% | Cpds 5% |
| ACE | 0.020 | 0.048 | 3.05 | 3 | 37 | 0.019 | 0.075 | 2.84 | 7 | 93 |
| ACHE | 0.024 | 0.038 | 3.74 | 8 | 80 | 0.019 | 0.107 | 4.11 | 22 | 200 |
| ADA | 0.020 | 0.000 | 0.00 | 0 | 20 | 0.018 | 0.000 | 0.00 | 0 | 49 |
| ALR2 | 0.098 | 0.028 | 3.74 | 2 | 21 | 0.071 | 0.154 | 6.80 | 9 | 52 |
| AMPC | 0.021 | 0.013 | 2.26 | 1 | 17 | 0.019 | 0.034 | 0.94 | 1 | 41 |
| AR | 0.161 | 0.157 | 11.96 | 19 | 59 | 0.108 | 0.268 | 7.83 | 31 | 147 |
| CDK2 | 0.087 | 0.117 | 9.70 | 14 | 43 | 0.063 | 0.190 | 5.24 | 19 | 108 |
| COMT | 0.000 | 0.091 | 4.35 | 1 | 10 | 0.000 | 0.182 | 5.44 | 3 | 24 |
| COX-1 | 0.154 | 0.113 | 11.82 | 6 | 19 | 0.102 | 0.250 | 7.17 | 9 | 47 |
| COX-2 | 0.322 | 0.234 | 18.03 | 154 | 275 | 0.193 | 0.397 | 10.14 | 216 | 686 |
| DHFR | 0.215 | 0.070 | 5.47 | 45 | 176 | 0.150 | 0.118 | 3.56 | 73 | 439 |
| EGFR | 0.048 | 0.038 | 3.26 | 31 | 330 | 0.036 | 0.071 | 2.19 | 52 | 824 |
| ER ago | 0.314 | 0.192 | 17.08 | 23 | 53 | 0.186 | 0.383 | 9.84 | 33 | 132 |
| ER antago | 0.059 | 0.110 | 8.90 | 7 | 30 | 0.040 | 0.173 | 5.08 | 10 | 75 |
| FGFR1 | 0.012 | 0.003 | 0.83 | 2 | 94 | 0.010 | 0.016 | 0.67 | 4 | 234 |
| FXA | 0.029 | 0.011 | 1.37 | 4 | 118 | 0.023 | 0.036 | 1.50 | 11 | 295 |
| GART | 0.108 | 0.000 | 0.00 | 0 | 19 | 0.087 | 0.005 | 1.00 | 2 | 46 |
| GPB | 0.113 | 0.026 | 2.87 | 3 | 44 | 0.081 | 0.101 | 4.22 | 11 | 110 |
| GR | 0.023 | 0.099 | 5.72 | 9 | 61 | 0.019 | 0.111 | 2.55 | 10 | 152 |
| HIVPR | 0.147 | 0.038 | 4.73 | 6 | 43 | 0.099 | 0.091 | 3.51 | 11 | 106 |
| HIVRT | 0.047 | 0.121 | 7.95 | 7 | 32 | 0.038 | 0.161 | 4.14 | 9 | 79 |
| HMGR | 0.015 | 0.035 | 2.79 | 2 | 31 | 0.012 | 0.049 | 1.14 | 2 | 76 |
| HSP90 | 0.039 | 0.000 | 0.00 | 0 | 21 | 0.032 | 0.004 | 0.54 | 1 | 51 |
| INHA | 0.079 | 0.191 | 12.04 | 21 | 68 | 0.051 | 0.257 | 6.50 | 28 | 168 |
| MR | 0.346 | 0.229 | 18.60 | 6 | 14 | 0.215 | 0.517 | 14.47 | 11 | 33 |
| NA | 0.019 | 0.000 | 0.00 | 0 | 39 | 0.018 | 0.000 | 0.00 | 0 | 97 |
| P38 | 0.031 | 0.012 | 1.54 | 14 | 192 | 0.026 | 0.049 | 2.29 | 52 | 480 |
| PARP | 0.114 | 0.071 | 4.24 | 3 | 28 | 0.080 | 0.091 | 3.39 | 6 | 70 |
| PDE5 | 0.047 | 0.009 | 1.68 | 3 | 42 | 0.037 | 0.043 | 1.81 | 8 | 104 |
| PNP | 0.011 | 0.000 | 0.00 | 0 | 22 | 0.009 | 0.000 | 0.00 | 0 | 55 |
| PPAR | 0.304 | 0.219 | 16.28 | 28 | 65 | 0.183 | 0.372 | 10.33 | 44 | 161 |
| PR | 0.012 | 0.009 | 1.80 | 1 | 22 | 0.010 | 0.027 | 1.47 | 2 | 54 |
| RXR | 0.653 | 0.330 | 26.47 | 11 | 16 | 0.362 | 0.620 | 14.81 | 15 | 39 |
| SAHH | 0.126 | 0.069 | 8.95 | 6 | 28 | 0.086 | 0.174 | 4.84 | 8 | 69 |
| SRC | 0.099 | 0.053 | 5.64 | 18 | 130 | 0.070 | 0.135 | 4.78 | 38 | 324 |
| THR | 0.129 | 0.097 | 7.57 | 11 | 51 | 0.091 | 0.149 | 3.87 | 14 | 127 |
| TK | 0.019 | 0.000 | 0.00 | 0 | 19 | 0.015 | 0.000 | 0.00 | 0 | 46 |
| TRP | 0.037 | 0.037 | 3.00 | 3 | 35 | 0.029 | 0.069 | 2.03 | 5 | 86 |
| VEGFR2 | 0.007 | 0.062 | 4.54 | 8 | 60 | 0.006 | 0.101 | 2.72 | 12 | 150 |
| Minimum | 0.000 | 0.000 | 0.00 | 0 | 10 | 0.000 | 0.000 | 0.00 | 0 | 24 |
| Maximum | 0.653 | 0.330 | 26.47 | 154 | 330 | 0.362 | 0.620 | 14.81 | 216 | 824 |
| Mean | 0.105 | 0.076 | 6.20 | 12 | 63 | 0.070 | 0.143 | 4.20 | 20 | 157 |
| Median | 0.048 | 0.048 | 4.24 | 6 | 39 | 0.038 | 0.101 | 3.51 | 10 | 97 |
Perhaps the first thing to note is the enrichment factor (after selecting the top 2% of the dataset) over all the targets varies from 0 to a maximum of 26 with a mean of 6. Where Enrichment factors were computed as follows:
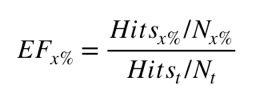
where Hitsx % is the number of active compounds in the top x % of the ranked dataset, Hitst is the total number of active compounds in the dataset, N x % is the number of compounds in the x % of the dataset and N t is the total number of compounds in the dataset. Unfortunately it is not possible to predict how much enrichment might be achieved.
Another way to look is to sort the data set by score and then plot number of ligands versus the number of active identified . For DHFR active ligands were identified among the highest scoring structures, but for GART the top 40 or so scoring ligands were inactives. The diagonal line gives an idea of the prevalence of hits with random picking.
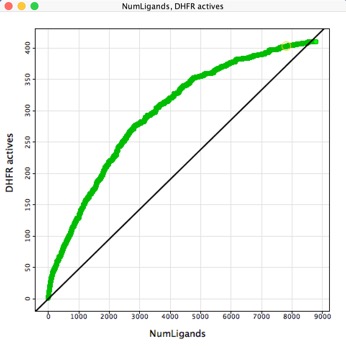
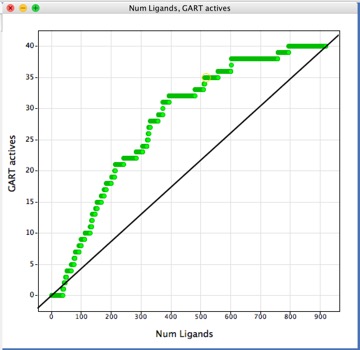
The objective for HTS Analysis is not to identify every active compound in the screening set, but rather to identify sufficient active series to support the active chemistry effort available, similarly the aim of virtual screening is not to identify every hit but rather to identify sufficient active series to support the active chemistry effort available. If we assume the percentage of true actives in the virtual library is 0.5% then the enrichment due to virtual screening might take it up to 3%. So for if you select 100 compounds for experimental determination one might expect 3 actives, if you want multiple series, (in case a series is lost due to off-target activity), you would probably want to evaluate 1000 compounds.
It is probably not wise to simply select the first 1000 compounds since it is likely that some chemotypes may be repeated, better to aim to select diverse chemotypes.
This might seem like a lot of compounds, but a back of the envelope calculation for the cost of a virtual screen is around $10,000 [taking into account hardware costs, licenses, maintenance and support, salaries], in addition you are probably going to be committing substantial biology and chemistry resources on any hits, so why would you want to penny pinch on the purchase of compounds?
Updated 14 Oct 2017