ENABLE-2 Incubator: Hit validation
ENABLE-2 offers evaluation of the antibiotic potential of novel compounds.
(i) Antibacterial activity (MIC) will be measured against selected Gram-negative and Gram-positive species to identify Hits (defined as compounds with wild-type whole cell activity on at least a species of interest).
(ii) Hits will be evaluated for in vitro cytotoxicity.
(iii) Limited hit expansion to explore SAR is possible if required by the programme and if ENABLE-2 resources are available.
Researchers at publicly funded universities and research institutes in Europe (including non-EU countries such as UK, Norway, Switzerland etc.) are eligible.
ENABLE-2 bacterial species of interest for Hit to Lead development
E.coli, K. pneumoniae, P. aeruginosa, A. baumannii, S. aureus, E. faecium.
UKRI funding to tackle antimicrobial resistance.
Transdisciplinary research to tackle antimicrobial resistance. Total fund £15,000,000.
You must be employed by a research organisation eligible to apply for UK Research and Innovation (UKRI) funding.
Your team and research project will bring new perspectives crossing Councils’ remits to understand and provide solutions to tackle AMR.
The full economic cost (FEC) of your project can be up to £3,000,000. UKRI will fund at 80% of the FEC.
The duration of the award is up to five years.
GARDP’s next webinar will take place on 7 June 2023
The latest Global Antibiotic Research & Development Partnership is on Project management in antimicrobial drug R&D.
Wed, Jun 7, 2023 12:30 PM - 2:00 PM BST
The webinar will feature the following speakers:
- Kristina Orrling, Programme Manager, Lygature, Netherlands
- Julie Miralves, R&D Portfolio and Planning Leader, Global Antibiotic R&D Partnership (GARDP), Switzerland
Register here https://register.gotowebinar.com/register/832555033649883487?source=network.
FDA Accepts Interim Analysis Plan for Ongoing Phase 2b Ibezapolstat Clinical Trial
FDA Accepts Interim Analysis Plan for Ongoing Phase 2b Ibezapolstat Clinical Trial and Acurx Announces Presentations at ECCMID 2023 Scientific Conference. This is an important step for Acurx, a great group of scientists to work with.
They will be presenting at the 33rd Annual European Congress of Clinical Microbiology and Infectious Disease (ECCMID) this month. Specifically, a scientific poster entitled "Novel pharmacology and susceptibility of ibezapolstat against C. difficile isolates with reduced susceptibility to C. difficile-directed antibiotics" will be presented by Dr. Kevin Garey, Professor and Chair, University of Houston College of Pharmacy and the Principal Investigator for microbiome aspects of our ibezapolstat clinical trial program.
Susceptibility testing in antibacterial drug R&D
This looks like an interesting webinar for all those interested in antibiotics research
Susceptibility testing in antibacterial drug R&D
Presentation 1: Pre-clinical antimicrobial susceptibility testing: considerations and challenges (Dee Shortridge):
- Steps for developing an in vitro susceptibility test for your lead compound
- Studies needed to characterize your compound
- Points to consider if your compound is not typical
Presentation 2: Pitfalls and opportunities of susceptibility testing in clinical trials of new antibiotics (Rafael Cantón):
- The introduction of new antimicrobials needs antimicrobial susceptibility testing to define their profile and the alignment with regulators.
- Susceptibility testing data are also used to define both clinical and PK/PD breakpoints.
- Moreover, they can be used to recognize wild type populations and anticipate emergence or resistance.
This is organised by the Global Antibiotic Research & Development Partnership (GARDP) https://gardp.org.
The Global Antibiotic Research & Development Partnership (GARDP) accelerates the development and access of treatments for drug-resistant infections. Together with private, public and non-profit partners, GARDP works to preserve the power of antibiotics for generations to come.
FIRST-IN-CLASS ANTIBIOTIC NOSO-502
It has been a real pleasure to be involved with the GNA NOW Consortium (https://amr-accelerator.eu/project/gna-now/) and I'm really delighted to share this news.
NOSOPHARM AND GNA NOW ANNOUNCE POSITIVE RESULTS FOR THE LATE PRECLINICAL DEVELOPMENT OF THE FIRST-IN-CLASS ANTIBIOTIC NOSO-502 An important milestone has been reached for the GNA NOW Consortium with the completion of the GLP toxicology studies for the NOSO-502 program. The results allow for the further development of the program to Phase 1.
Full details are here https://www.lygature.org/news/gna-now-consortium-announces-positive-results-late-preclinical-development-first-class.
The NOSO-502 program received a unanimous recommendation from the internal and external experts of the GNA NOW Consortium to start preparing for clinical trials. This is of particular importance as novel classes of antibiotics with efficacy against the WHO critical priority Gram-negative pathogens are very rare. No novel class of antibiotics with efficacy against these pathogens has been introduced into clinical use for more than 40 years. Furthermore, according to a very recent and comprehensive analysis of the antibacterial drug pipeline, there is no first-in-class Gram-negative antibiotic with a novel target or a novel mode of action in clinical development. If successful, the introduction into the clinical use of NOSO-502 will give a new option to the physician for the treatment of patients suffering from life-threatening bacterial infections, avoiding a therapeutic dead-end. This way, NOSO-502 will strengthen the therapeutic arsenal against Gram-negative infections.
Fantastic news to all involved in the program and great news for patients.
NICE reaches important milestone in the UK’s efforts to tackle antimicrobial resistance.
Two new antimicrobial drugs - cefiderocol and ceftazidime–avibactam - are close to becoming the first to be made available as part of the UK’s innovative subscription-style payment model after NICE published draft guidance estimating their value to the NHS.
Cefiderocol is a cephalosporin antibiotic that is coupled to a siderophore that binds to iron and aids cell entry.
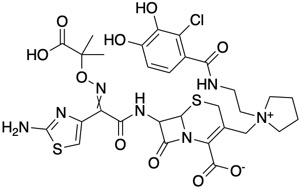
Ceftazidime–avibactam is a fixed-dose combination medication composed of ceftazidime, a cephalosporin antibiotic, and avibactam, a β-lactamase inhibitor. Bacterial resistance to cephalosporins is often due to bacterial production of β-lactamase enzymes that deactivate these antibiotics. Avibactam inhibits bacterial β-lactamases.
Investment in new antimicrobials, especially those that target multi-drug-resistant pathogens, is not commercially attractive because they are subject to strict controls to restrict their use to slow the development of resistance. This means sales could be low. The new payment method overcomes this by ensuring a fixed annual fee is paid to the company regardless of how many prescriptions are issued.
On the Antibacterial Action of Cultures of a Penicillium
May 10th 1929 is a very important day in drug discovery research, it was on this day that Alexander Fleming submitted his paper entitled "On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ" Br J Exp Pathol. 1929 Jun; 10(3): 226–236.

Over 90 years later this chance discovery still has a major impact on health today. Whilst isolation proved too challenging for Fleming he sent his Penicillium mold to anyone who requested it in hopes that they might isolate penicillin. It was only in 1940 that Howard Florey and team published the isolation and purification of Penicillin. Penicillin as a chemotherapeutic agent. Lancet. 1940;236:226–8. 10.1016/S0140-6736(01)08728-1.

The basic structure of the beta-lactam ring is shown below.
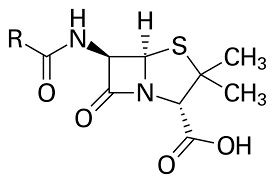
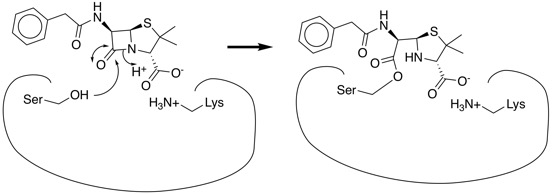
The beta lactam antibiotics (Penicillin and Cephalosporins) are a very well studied class of therapeutic agent, the mechanism of action is the inhibition of cell wall synthesis. Penicillin inhibits the formation of peptidoglycan cross-links in the bacterial cell wall; this is achieved through reaction of the β-lactam ring of penicillin to the enzyme DD-transpeptidase. As a consequence, DD-transpeptidase cannot catalyze formation of the cell wall cross-links.
Funding for novel antibiotic development
Registration is open for a GNA NOW #webinar taking place on Wednesday 12 May from 10:00-11:00 (CEST).
The GNA NOW Consortium is looking for a novel antibiotic compound to progress to a clinical candidate. To find out more about what the Consortium is looking for, what’s in it for you as a researcher, and how to submit a proposal, make sure not to miss this webinar! The session will feature presentations by Kristina Orrling from Lygature and Eric Bacqué of Evotec.
Register for free https://www.lygature.org/registration-gna-now-open-call-webinar
Funded by the @Innovative Medicines Initiative (IMI)
New GARDP webinars

25 February: 'From discovery to IND: Roadmap to a successful antibacterial project' with Patricia Bradford and Alita Miller, moderated by Michael Mourez. Register here: https://attendee.gotowebinar.com/register/8974462807057685772?source=spark
4 March: 'Learning from COVID-19 to tackle the silent pandemic of antibiotic resistance' with Marc Mendelson, Joanne Liu and Manica Balasegaram. Register here: https://attendee.gotowebinar.com/register/3151890131955239691?source=spark
24 March: 'Discovering and developing new treatments for tuberculosis' with Nader Fotouhi, moderated by Lydia Nakiyingi. Register here: https://attendee.gotowebinar.com/register/394450209091477264?source=spark
As always, keep an eye on our website https://revive.gardp.org/webinars to find new webinar announcements and recordings of previous webinars.
Open Source Antibiotics
Open Source Antibiotics is a consortium of researchers interested in open ways to discover and develop new, inexpensive medicines for bacterial infections.
There are already a couple of projects Mur Ligase and a series of Diarylimidazoles with unknown mechanism. Well worth a read.
All the structures of the molecules on the project are openly available in a spreadsheet https://docs.google.com/spreadsheets/d/168-a1_l51Nfbms67eG8zU8p-EhEtEO26FUzRInbu7fY/edit#gid=2078630269 feel free to have a browse.
New REVIVE Antimicrobial Encyclopaedia
Often when moving into a new therapeutic area it takes a while to pick up all the terms and acronyms, and antimicrobial research has some of it's own, not sure what a MIC is then head over to the REVIVE Antimicrobial Encyclopaedia and search.
The REVIVE Antimicrobial Encyclopaedia includes definitions of terms from the field of antimicrobials including ‘Active Pharmaceutical Ingredient’, ‘Bacterial efflux’ and ‘Minimum Inhibitory Concentration’. Each term has links for users to find more information on the subject and wherever available there are also links to REVIVE content such as webinar recordings and Antimicrobial Viewpoints on the subject. Some terms also include bespoke explanatory videos with clear diagrams featuring REVIVE experts.
REVIVE is a space for everybody with an interest in antimicrobial R&D. The Global Antibiotic Research and Development Partnership (GARDP) is a not-for-profit organization developing new treatments for drug-resistant infections that pose the greatest threat to health. We were created to ensure that everyone who needs antibiotics receives effective and affordable treatment, no matter where they live. We aim to develop five new treatments by 2025 to fight drug-resistant infections, focusing on sexually transmitted infections, sepsis in newborns and infections in hospitalized adults and children.
Open Source Antibiotics
I just thought I'd share this email
Dear Friend of Open Source Antibiotics (OSA),
It's been a busy few months in OSA. Recent activity is being captured in weekly public Zoom meetings (every Friday at 2pm London at https://ucl.zoom.us/j/92800004715), and you can see the details in the recordings of those meetings (like this one) and the associated "Github Issues" (like this one).
But as part of those discussions we were wondering about the best way to update everyone quickly. While OSA uses Twitter, there is no good substitute for a short email. So this is the first, short OSA news email. Three points:
- We have guessed you're interested in receiving occasional emails about OSA. If you're not, just email us back to say you'd like to opt out. Nobody likes spam.
- Please forward this to anyone you think might be interested in antibiotics or drug discovery or open science. As an open project, everything is in the public domain, and everyone is welcome.
- To keep things short, each news email has a limit of three items. If you're interested in learning more, then each project has a wiki (current project status, like this) and the Issue Tracker (current To Do list and discussion, like this).
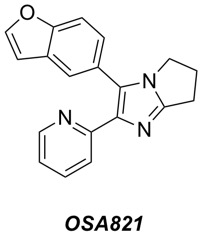
- We have confirmed the activity vs MRSA of the diarylimidazoles (exemplar compound OSA821 shown below), originally discovered and explored by Alvaro Lorente and Bill Zuercher at UNC Chapel Hill. A new potency screen is being performed this week at UCL by Paul Stapleton, and includes about 30 compounds that have been donated to the project via Ben Perry (DNDi). This time our potency assay will include a parallel screen of select compounds vs VRE, to see if there is activity vs other high priority Gram +ves.
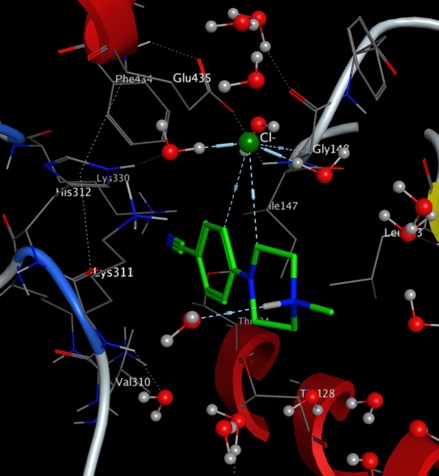
- A key aim of the project is to solve the rapid clearance of the known actives. New data from Sue Charman's lab gave clues as to which compounds to investigate next, and we are finalising negotiations for some pro bono work from a UK company towards identification of possible metabolites.
- The mechanism of action of these compounds is unknown, but Lee Graves's lab at UNC are in the middle of some MIBs experiments that we hope will reveal, by the end of October, some key new insights into how the compounds work.

You can read more about the Open Source Antibiotics on GitHub https://github.com/opensourceantibiotics.
There are currently two projects MurLigase and DiarylImidazoles, everything is in the open and anyone can contribute.
Why not swing by and have a browse.
GARDP: Bringing new treatments for drug-resistant infections to all who need them
This webinar provided an overview and update on GARDP’s efforts to bring new antibiotic treatments for drug-resistant infections to all who need them.
The following topics were presented:
- Antibiotic resistance and the GARDP response
- Tackling the growing threat of hospital infections
- Developing new treatments for neonatal sepsis
Discovery of novel antibiotic Halicin using deep learning
A recent paper has caught a lot of attention recently "A Deep Learning Approach to Antibiotic Discovery" DOI from Regina Barzilay's group at MIT. They used a deep neural network model to predict growth inhibition of Escherichia coli using a collection of 2,335 molecules, the molecules were described using Morgan fingerprints, computed using RDKit, for each molecule using a radius of 2 and 2048-bit fingerprint vectors. Using this methodology they identified the known c-Jun N-terminal kinase inhibitor SU3327 which they renamed Halicin. A quick search using MolSeeker allowed identification of the structure and inChiKey.
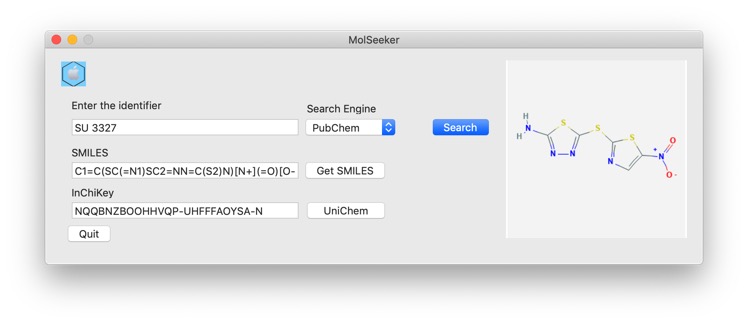
A search of UniChem using the InChikey NQQBNZBOOHHVQP-UHFFFAOYSA-N identified a number of other identifiers in different databases.
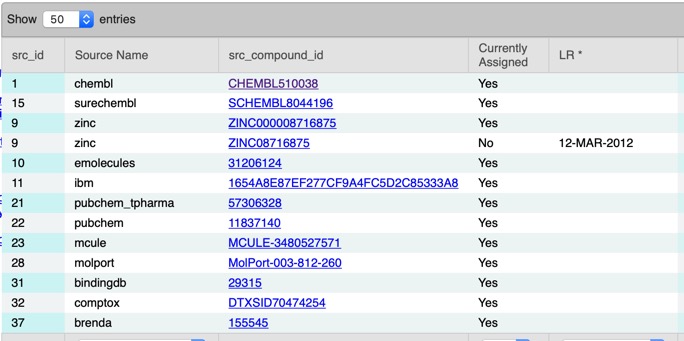
Including a link to the ChEMBL entry CHEMBL510038 giving the biological data 0.7 nM Inhibition of c-Jun N-terminal kinase by time-resolved FRET assay, and links to the original 2009 publication DOI describing the c-JNK SAR. The compound has a rat half-life of 0.45 h. There is another publication that might be of interest describing "Discovery of 2-(5-nitrothiazol-2-ylthio)benzo[d]thiazoles as novel c-Jun N-terminal kinase inhibitors" DOI.
Certainly an interesting approach, I suspect the nitrothiazole functionality would set off a few structural alerts but there are certainly of plenty of similar compounds commercially available that would allow exploration of the SAR without too much investment in resources.
All code and data is available on GitHub and there is also a website where you can test your own molecules http://chemprop.csail.mit.edu.
Call for Open Source Antibiotics Fragment Contributions
What we have: Fragment hits from an initial screen against MurE and MurD, performed at Diamond screening facility, and a platform to screen additional fragment libraries or follow-up compounds.
What we need: Additional chemical matter for screening. The Diamond screening platform is high-throughput and we would ideally be able to take full advantage of this.
Full details are on the Open Source Antibiotics website
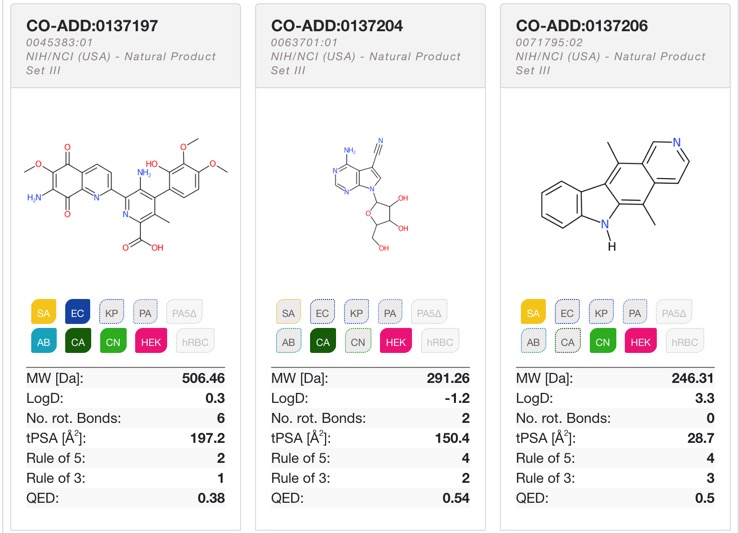
Open-access Antimicrobial Screening Database
I just got news of the first public release of CO-ADD screening data
CO-ADD is a non-for-profit initiative led by academics at The University of Queensland. Our goal is to screen compound for antimicrobial activity for academic research groups and generate a public knowledge database for the development of novel agents for the treatment of microbial infections. The knowledge base contains chemical structures and antimicrobial activity data from CO-ADD’s screening, made publicly available by the academic research groups, with more data to be released over time.
The database is available here.
OpenSource Antibiotics
I just thought I’d highlight a new project I’m involved with.
Open Source Antibiotics (https://github.com/opensourceantibiotics) is intended to be a platform for a collaborative effort towards antibiotic discovery.
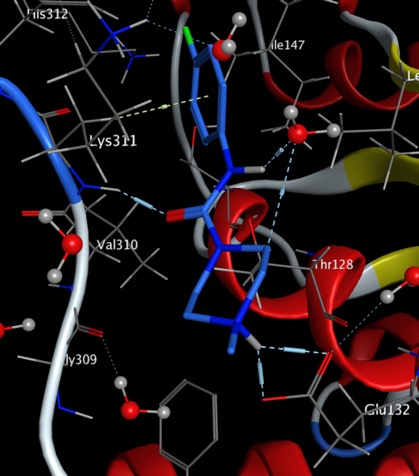
The first projects have been initiated
Mur Ligase (https://github.com/opensourceantibiotics/murligase) and the background to these exciting targets can be found on the wiki page.
https://github.com/opensourceantibiotics/murligase/wiki
This also provides details of the first two fragment screens.
MurD https://github.com/opensourceantibiotics/murligase/wiki/MurD-fragment-screen
and
MurE https://github.com/opensourceantibiotics/murligase/wiki/MurE-fragment-screen
What we want now is for people to join in and suggest the next round of fragments that should be screened. Ideally these should be commercially available but if people want to design, make and submit their own fragments we would be happy to screen them.
If you feel appropriate, we would appreciate any publicity on this exciting new project
Causes of death over 100 years
The UK Office of National Statistics has produced a fascinating interactive plot of the causes of death in the UK over the last 100 years.
I've captured a screenshot of the plots but I'd urge to go and have a look at the interactive plot on the website http://visual.ons.gov.uk/causes-of-death-over-100-years/.
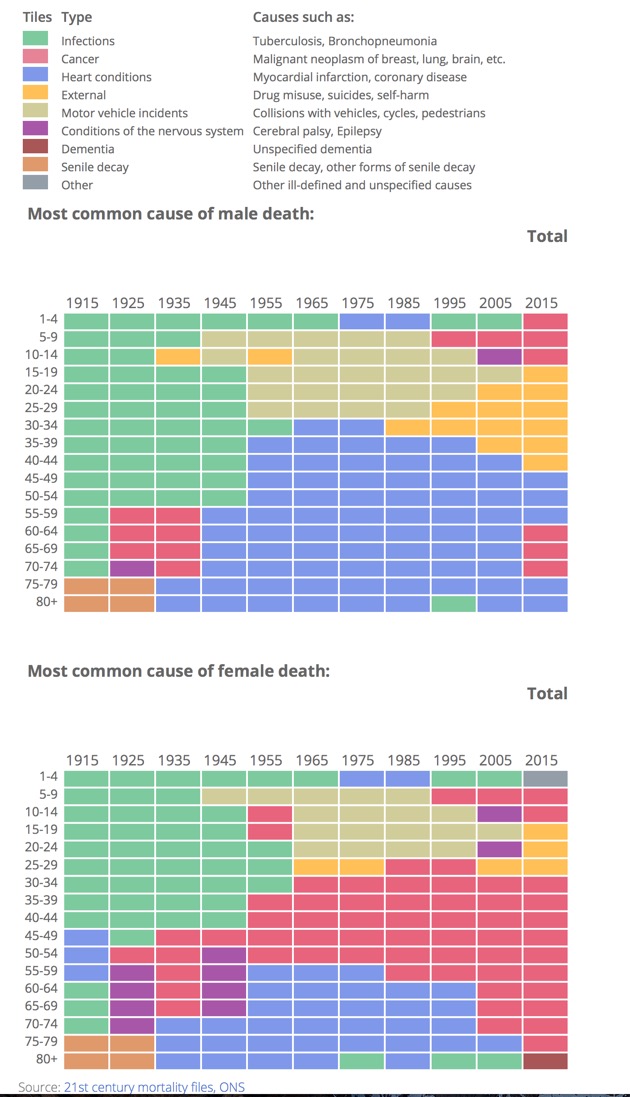
What is very apparent is the impact the introduction of antibiotics had in the late 1940's, and the introduction of mass vaccinations, deaths due to infections have been virtually eliminated.
In men heart disease remains the major killer whilst in women it is breast cancer. Sadly among the young it looks like mental health issues are a major concern.
ChEMBL_23 released
ChEMBL_23 has been released, it was prepared on 1st May 2017 and contains:
- 2,101,843 compound records
- 1,735,442 compounds (of which 1,727,112 have mol files)
- 14,675,320 activities
- 1,302,147 assays
- 11,538 targets
- 67,722 source documents
Data can be downloaded from the ChEMBL ftp site: ftp://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/releases/chembl_23
Also includes
Deposited Data Sets
CO-ADD, The Community for Open Antimicrobial Drug Discovery, is a global open-access screening initiative launched in February 2015 to uncover significant and rich chemical diversity held outside of corporate screening collections. CO-ADD provides unencumbered free antimicrobial screening for any interested academic researcher. CO-ADD has been recognised as a novel approach in the fight against superbugs by the Wellcome Trust, who have provided funding through their Strategic Awards initiative. Open Source Malaria (OSM) is aimed at finding new medicines for malaria using open source drug discovery, where all data and ideas are freely shared, there are no barriers to participation, and no restriction by patents. The initial set of deposited data from the CO-ADD project consists of OSM compounds screened in CO-ADD assays (DOI = 10.6019/CHEMBL3832881).
Modelled on the Malaria Box, the MMV Pathogen Box contains 400 diverse, drug-like molecules active against neglected diseases of interest and is available free of charge (http://www.pathogenbox.org). The Pathogen Box compounds are supplied in 96-well plates, containing 10 uL of a 10mM dimethyl sulfoxide (DMSO) solution of each compound. Upon request, researchers around the world will receive a Pathogen Box of molecules to help catalyse neglected disease drug discovery. In return, researchers are asked to share any data generated in the public domain within 2 years, creating an open and collaborative forum for neglected diseases drug research. The initial set of assay data provided by MMV has now been included in ChEMBL (DOI = 10.6019/CHEMBL3832761).
WHO publishes list of bacteria for which new antibiotics are urgently needed
The World Health Organisation has published a list of the top 12 bacteria for which antibiotics are urgently needed in an effort to focus research.
The list highlights in particular the threat of gram-negative bacteria that are resistant to multiple antibiotics. These bacteria have built-in abilities to find new ways to resist treatment and can pass along genetic material that allows other bacteria to become drug-resistant as well.
WHO priority pathogens list for R&D of new antibiotics
Priority 1: CRITICAL
- Acinetobacter baumannii, carbapenem-resistant
- Pseudomonas aeruginosa, carbapenem-resistant
- Enterobacteriaceae, carbapenem-resistant, ESBL-producing
Priority 2: HIGH
- Enterococcus faecium, vancomycin-resistant
- Staphylococcus aureus, methicillin-resistant, vancomycin-intermediate and resistant
- Helicobacter pylori, clarithromycin-resistant
- Campylobacter spp., fluoroquinolone-resistant
- Salmonellae, fluoroquinolone-resistant
- Neisseria gonorrhoeae, cephalosporin-resistant, fluoroquinolone-resistant
Priority 3: MEDIUM
- Streptococcus pneumoniae, penicillin-non-susceptible
- Haemophilus influenzae, ampicillin-resistant
- Shigella spp., fluoroquinolone-resistant
Update
The Community for Open Antibiotic Drug Discovery (CO-ADD) screen compounds for antimicrobial activity for academic research groups for free. The screening includes the top 5 pathogens listed in the WHO priority list, as well as the fungi C. neoformans and C. albicans. Details on how to send compounds are here. All they require is 1mg (or 50uL at 10 mg/mL) of pure compound which will be used for primary screening, hit confirmation, and if active will be used for a broader antimicrobial screening, cytotoxicity and a check for its purity.