Structure-based design.
Structure-based design relies on having knowledge of the three dimensional structure of the molecular target for drug, the principle means of deriving the 3D structure are X-ray crystallography and solution NMR, alternatively it may be possible to build a homology model based on a related protein.
The Quality of the 3D structure is critical
This can not be understated.
We tend to believe that PDB files are a message from God (Derek Lowe, In The Pipeline).
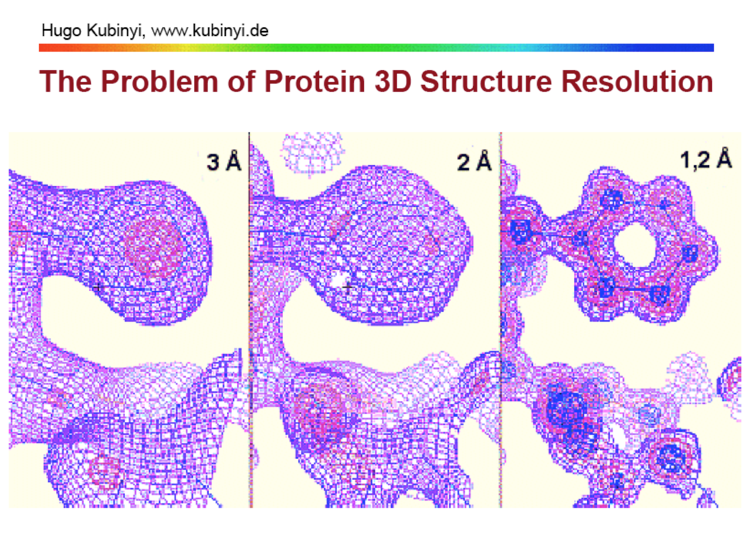
The quality of an X-ray crystal structure can vary and it is useful to understand how the model is derived from the electron density maps. The crystallographer builds a 3D model of the protein attempts to fit it to the electron density then uses the built structure to generate an electron density map, they then use this to derive a difference map to show where the modelled structure does not match the experimental density map. The modelled structure is then adjusted and the process repeated until they have a refined structure.
As the image below highlights the resolution can have a significant impact on the quality of the eventual structure.
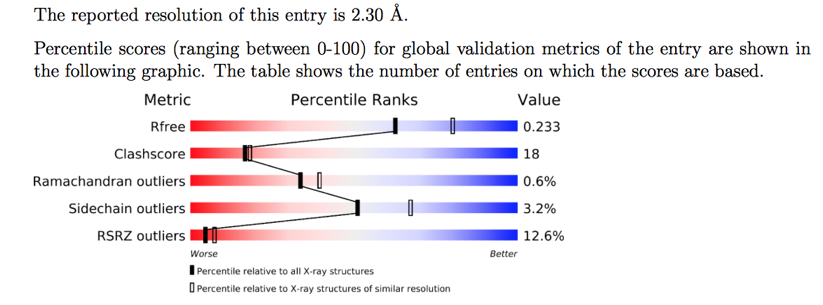
If you are downloading a PDB structure the page for each entry provides a lot of very useful information, giving the resolution and a graphical display of various parameters (red=Poor, Blue=Good).
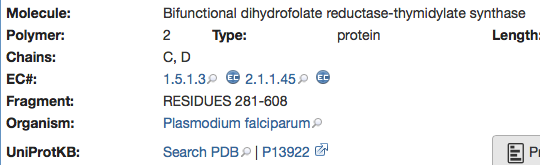
The Molecular Description provides information about the protein but also indicates which residues are in the crystal structure, many crystal structures may have been modified to aid crystallisation.
This information is also contained within the header of the PDB file you can download.
Once you have downloaded the PDB file you can open it in a text editor to view the header information or in a molecular modelling package to visualise the protein. Before you start any work on the protein however it is important to examine the structure for potential errors, as shown in the screenshot below.
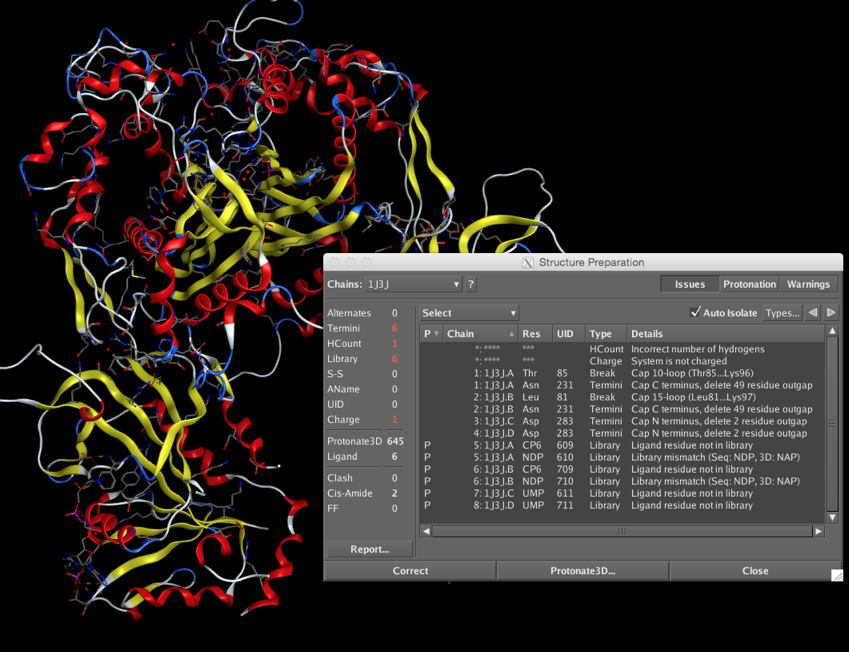
Some of the potential issues are :-
- Alternates, Residues with alternate locations and/or ambiguous sequence identities (choose highest occupancy)
- Termini, Protein chain C- or N-termini which need to be charged or capped, or if DNA the terminal PO4 may only have three oxygens bonded to the phosphorous and an additional oxygen needs to be added. Sometimes loops are very disordered and appear as a breaks in the chain, it may be possible to use a loop library to model a replacement.
- Hydrogens, usually not visible and so need to be added/checked, particularly check hydrogens on heteroatoms, especially active site residues where the local environment may influence pKa
- Ligand, Novel ligands in particular need checking to confirm atoms and bond orders are correct
- Conformation, check that torsions are reasonable and there are no clashes.
- Charge, It with worth checking the charge on all ionisable groups.
- It can be difficult to be certain of the position of nitrogens in His or the primary amide in Asn, Gln.
Binding Site Identification
In many cases the X-ray structure (or a related protein) may contain a bound ligand, in which case it should be straight-forward to define the binding site, however if no ligand is present (apoprotein) or there are unoccupied allosteric site it may be possible to use computational tools to identify potential binding sites. Examples of such computational tools include Sitefind, SiteHound, and PDBinder.
Worth Reading, In silico prediction of binding sites on proteins, CURRENT MEDICINAL CHEMISTRY 17(15):1550-62 · FEBRUARY 2010, DOI.
How well does my ligand bind?
Prediction of binding affinity is an extremely challenging problem, the approaches can be separated into three categories, Empirical scoring functions, Knowledge-based, and Forcefield methods.
Knowledge-based scoring functions. Here, binding affinity is considered as a sum of the ligand and protein atoms interactions. As the empirical scoring functions, these potentials are derived from experimental structures, where interatomic distances are converted into distance-dependent interactions free energies. These functions are designed to reproduce experimental structures rather than binding energies. Due to its simplicity, knowledge-based scoring functions permit rapid screening of large compound databases
Empirical scoring functions. The binding free energy is estimated based on weighted structural parameters by fitting the scoring functions to experimental determined binding constants of a set of complexes. These scoring functions may be potentially biased by the selected training set of ligand-protein complexes. The advantage of these functions is that their terms, although similar to force-field functions, are orders of magnitude easier to evaluate. More complex functions attempt to addresses solvation and desolvation effects but since these effects are poorly understood they provide only incomplete descriptions of these effects on protein-ligand binding.
Force-field scoring functions. Make use of classical molecular mechanics for energy function calculations. The binding free energy of protein-ligand complexes are estimated by the sum of van der Waals (by Lennard-Jones potential function) and electrostatics interactions. Solvation is considered as a distance-dependent dielectric function. Non-polar contributions are assumed to be proportional to the solvent-accessible surface area. Non-bonded interactions are treated with the introduction of a cut-off distance. The method requires energy minimization of the complex prior to energy evaluations. Various force-field scoring functions are based on different sets of parameters.
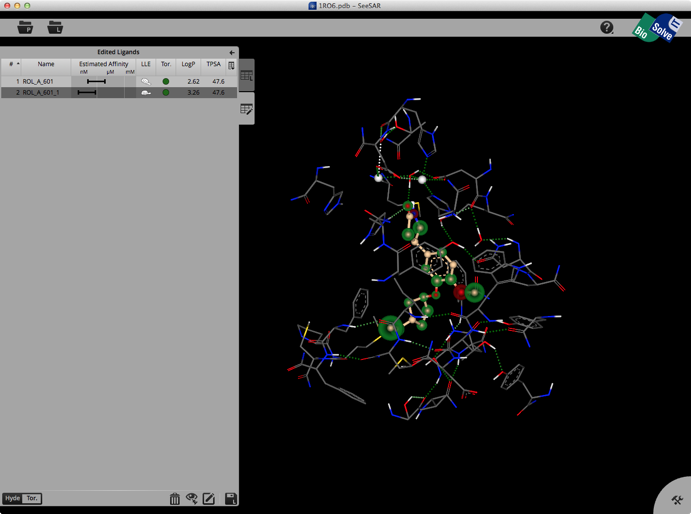
A particular interesting tool to aid medicinal chemists in structure-based design is SeeSAR an interactive tool for designing/improving ligands for drug discovery. The structure display is pretty intuitive, green is a favourable interaction, red unfavourable, with the size of the coloured spheres indicating the magnitude of the interaction. It also gives an estimated affinity and LLE.
Worth Reading,
Assessment of Programs for Ligand Binding Affinity Prediction, Journal of Computational Chemistry DOI
Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database, Journal of Computational Chemistry Volume 32, Issue 4, pages 742–755, March 2011. DOI.
A consistent description of HYdrogen bond and DEhydration energies in protein–ligand complexes: methods behind the HYDE scoring function Journal of Computer-Aided Molecular Design
January 2013, Volume 27, Issue 1, pp 15-29 DOI.
Last update 13 March 2020