COVID-19 Registered Trials
There are now a number of clinical trials underway and this review by The Centre for Evidence-Based Medicine provides an excellent summary of the trials that are taking place. They describe proposed pharmacological interventions and their mechanisms, when known, but unfortunately don't give the chemical structures.
Updated
I've also now included a few other structures that people have sent to me.
Here is the workflow I use to get the structures and access more information about the compounds.
Create a text file with all the structures mentioned
ASC09
Azvudine
Azithromycin
Baloxavir
Carriomycin
Chloroquine
cobicistat
Danoprevir
Darunavir
Dihydroartemisinin
Favipiravir
Fingolimod
hydroxychloroquine
Jakotinib
Leflunomide
Lopinavir
marboxil
Methylprednisolone
oseltamivir
piperaquine
Remdesivir
ribavirin
ritonavir
Ruxolitinib
Suramin
Thalidomide
thymosin
Triazavirin
Umifenovir
Now read the text file into Vortex
The use a Name to Structure script to use a web service to get the structures, in this case I used ChemSpider. Now generate the InChiKey from the structures.
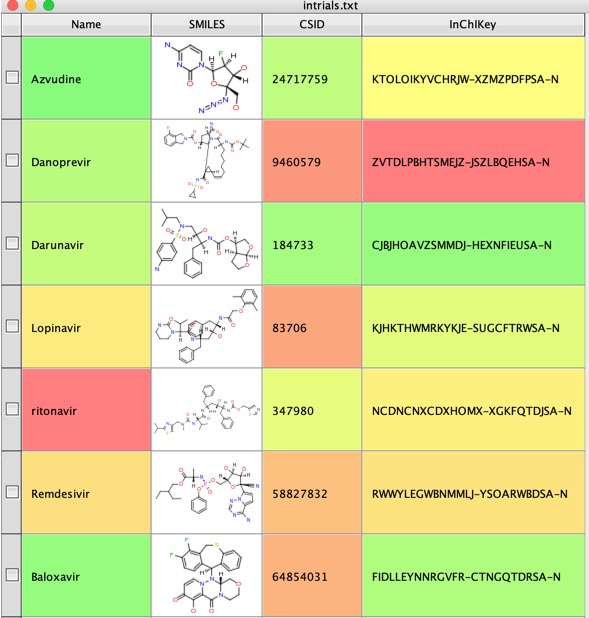
We can now use the InChiKey to search UniChem using another Vortex script to get identifiers for the molecule from various databases.
UniChem efficiently produces cross-references between chemical structure identifiers from different databases

We can then use the identifiers to search the various databases for more information
I've been asked if I could provide the structures for download
Here it is in SDF file format http://cambridgemedchemconsulting.com/news/files/COVID19/coviddata.sdf.zip
And in SMILES format http://cambridgemedchemconsulting.com/news/files/COVID19/forpost.smi.zip.