Alphafold in Opentargets
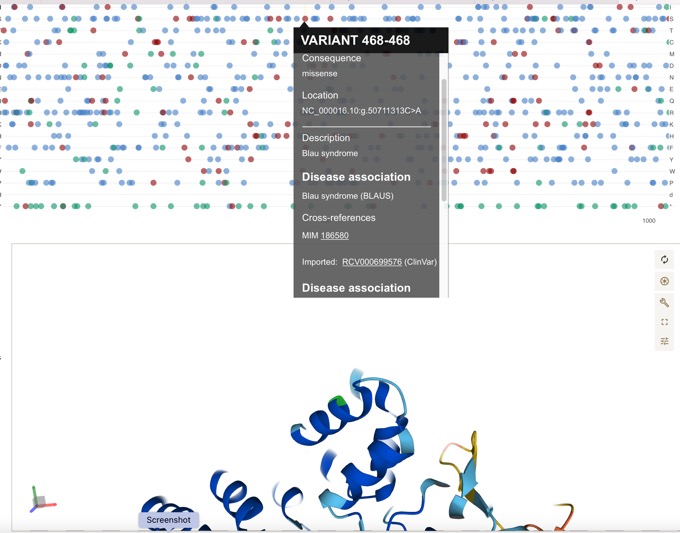
Thanks to fantastic work from the folks at UniProt, the Open Targets Platform target profile pages now feature DeepMind’s AlphaFold data.
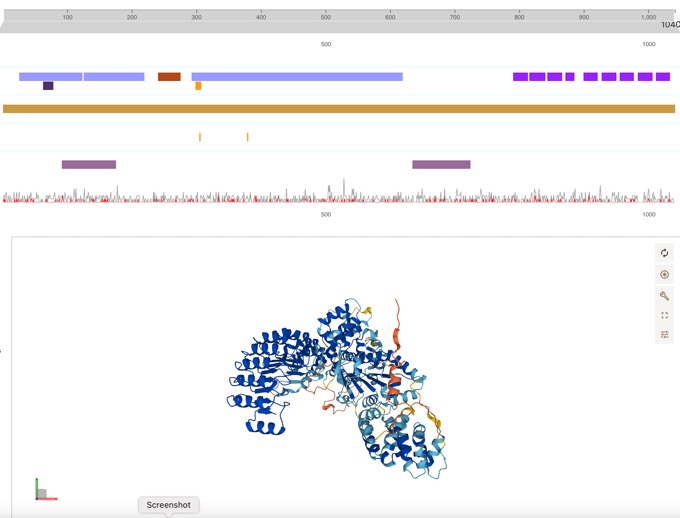
Can be easily linked to disease associations.
Open Targets Platform 21.06 has been released!
Open Targets Platform 21.06 has been released
The Open Targets Platform is a comprehensive tool that supports systematic identification and prioritisation of potential therapeutic drug targets. By integrating publicly available datasets including data generated by the Open Targets consortium, the Platform builds and scores target-disease associations to assist in drug target identification and prioritisation. It also integrates relevant annotation information about targets, diseases, phenotypes, and drugs, as well as their most relevant relationships.
Currently there are:-
Targets 60,606
Diseases 18,507
Drugs 13,185
Evidence strings 13,267,236
Associations 11,755,362
Open Targets Platform: rebuilt, redesigned, reimagined
The Open Targets Platform has been updated.
- The new design of the Platform improves user experience and access to information. It has been reimagined to facilitate the building of new therapeutic hypotheses;
- New features include data on binary molecular interactions, and black box warnings;
- A complete refactoring of the codebase enables rapid development and new deployment strategies.
Chemical Probes
A really interesting review of the Chemical Probes portal
2020 was the first year of visible activity on the Chemical Probes Portal since 2017, with 115 probes added and over 500 compounds now included on the Portal. To celebrate, we’re highlighting ten of the best probes added to the Portal and evaluated by our Scientific Advisory Board in 2020. These probes are selective, potent, cell-active molecules that are rated four stars for use in cells and target new proteins or have new mechanisms of action. They include probes for previously ‘undruggable’ cancer targets, compounds that target GPCRs, epigenetic modulators and PROTACs.
Full details are here https://www.chemicalprobes.org/news/2020s-top-probes.
Open Targets Platform beta test
If you have a little time why not drop by the Open Targets website and give the beta test version and give them some feedback. Full details are in the blog post.
The beta version makes the most of the data from the recent 21.02 release and you can also interrogate the data using the brand new GraphQL API.
In particular, this version features:
- Redesigned evidence pages
- Updated drug profile pages
- More complete disease profile pages
Open Targets is a public-private partnership that uses human genetics and genomics data for systematic drug target identification and prioritisation. The current focus is on oncology, immunology and neurodegeneration.
Generating and interpreting the data required to identify a good drug target demands a diverse set of skills, backgrounds, evidence types and technologies, which do not exist today in any single entity. Open Targets brings together expertise from seven complementary institutions to systematically identify and prioritise targets from which safe and effective medicines can be developed.
OpenTargets updated
Just had an email about the latest Open Targets Platform release - 19.11.
In this release there is data on
- 27,069 targets
- 13,579 diseases
- 8.91 million pieces of evidence
- 6.33 million associations between targets and diseases
History of rare diseases and their genetic causes - a data driven approach
One of the advantages of being a consultant is that I can feel free to contribute to projects that I find interesting. So as well as working with a couple of Open-Source drug discovery projects (e.g. Open Source Antibiotics I can also follow a couple of rare disease programs.
This publication looks very useful History of rare diseases and their genetic causes - a data driven approach.
This dataset provides information about monogenic, rare diseases with a known genetic cause supplemented with manually extracted provenance of both the disease and the discovery of the underlying genetic cause of the disease.
More details of how the dataset was constructed.
We collected 4166 rare monogenic diseases according to their OMIM identifier, linked them to 3163 causative genes which are annotated with Ensembl identifiers and HGNC symbols. The PubMed identifier of the scientific publication, which for the first time describes the rare disease, and the publication which found the gene causing this disease were added using information from OMIM, Wikipedia, Google Scholar, Whonamedit, and PubMed. The data is available as a spreadsheet and as RDF in a semantic model modified from DisGeNET.
A very interesting read.
Open Targets Updated
The latest Open Targets Platform 19.09 has been released. The latest release contains
27,024 targets 10,474 diseases 3.33 million pieces of evidence 7.78 million associations between targets and diseases
In addition, a number of Target Enabling Packages (TEP) provided by Structural Genomics Consortium have been included, there are more details here. Several new chemical probes have also been included.
Open Targets Platform: release 19.06 is out
The latest update of the Open Targets Platform, release 19.06 is available.
This update includes
Target safety information
As a follow-up to the safety data in Open Targets Platform release 19.04, now has more targets with known safety effects and safety risk information, including TBXA2R and JAK2.
TEPs and chemical probes
In this release, they've included the latest Target Enabling Packages (TEPs) for GALT, GALK1 and MLLT1. Also added more chemical probes, small-molecule modulators of a protein’s function that can be used in cell-based or animal studies.
Target-disease associations
A new release always means new evidence available for novel target-disease associations.
Cathepsin C inhibitor chemical probe
As part of the Boehringer Ingelheim's efforts to foster innovation, they are share selected molecules with the scientific community all for free. The opnme portal gives access to a range of novel ligands. The latest addition is BI-9740
BI-9740 is a very potent and highly selective inhibitor of the enzymatic activity of Cathepsin C. It blocks human CatC in vitro with an IC50 of 1.8 nM and shows > 1500x selectivity versus the related proteases Cathepsin B, F, H, K, L and S. BI-9740 displays no activity against 34 unrelated proteases from different classes up to a concentration of 10 µM.
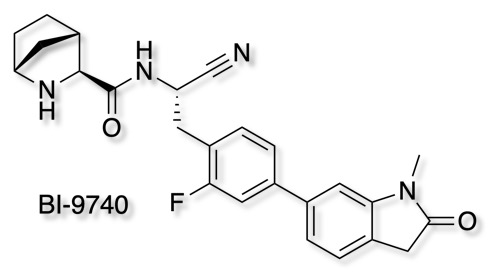
Chemical probes are absolutely essential for target validation and it is great to see so many high quality tools being made available.
Promises, promises, and precision medicine
A very interesting commentary on the impact (or lack of) genomics has had on human healthcare. J Clin Invest. 2019
The promises of precision medicine are to dramatically change patient care via individually tailored therapies and, as a result, to prevent disease, improve survival, and extend healthspan.
However, nearly two decades after the first predictions of dramatic success, we find no impact of the human genome project on the population’s life expectancy or any other public health measure, notwithstanding the vast resources that have been directed at genomics. Exaggerated expectations of how large an impact on disease would be found for genes have been paralleled by unrealistic timelines for success, yet the promotion of precision medicine continues unabated.
In light of the limitations of the precision medicine narrative, it is urgent that the biomedical research community reconsider its ongoing obsession with the human genome and reassess its research priorities including funding to more closely align with the health needs of our nation. We do not lack for pressing public health problems. We must counter the toll of obesity, inactivity, and diabetes; we need to address the mental health problems that lead to distress and violence; we cannot stand by while a terrible opiate epidemic ravages our country; we have to prepare conscientiously for the next influenza pandemic; we have a responsibility to prevent the ongoing contamination of our air, food, and water. Topics such as these have taken a back seat to the investment of the NIH and of many research universities in a human genome–driven research agenda that has done little to solve these problems, but has offered us promises and more promises.
The human genome project was undoubtedly a magnificent achievement, but has the investment in genomics delivered?
There is an extended discussion on In the Pipeline https://blogs.sciencemag.org/pipeline/archives/2019/01/31/precision-medicine-real-soon-now.
Open Targets Genetics
As mentioned on the Target Validation page, Open Targets is a public-private partnership that uses human genetics and genomics data for systematic drug target identification and prioritisation. The current focus is on oncology, immunology and neurodegeneration. An extension to this will be launched on 18 October 2018.
Open Targets Genetics is a new portal developed by Open Targets, an innovative partnership that brings together expertise from six complementary institutions to systematically identify and prioritise targets from which safe and effective medicines can be developed. The Portal offers three unique features to help you discover new associations between genes, variants, and traits, Gene2Variant analysis pipeline, Fine mapping/ credible set analysis, UK Biobank + GWAS Catalog integration.
More details https://genetics.opentargets.org/docs/open-targets-genetics-infographic.pdf
Open Targets Platform Update
Just announced. Open Targets Platform release - 18.08.
In this release there is now data on:
- 21,149 targets
- 10,101 diseases
- 6.5 million pieces of evidence
- 2.9 million associations between targets and diseases
The Open Targets Platform is a comprehensive and robust data integration for access to and visualisation of potential drug targets associated with disease. It brings together multiple data types and aims to assist users to identify and prioritise targets for further investigation. A drug target can be a protein, protein complex or RNA molecule and it’s displayed by its gene name from the Human Gene Nomenclature Committee, HGNC. We integrate all the evidence to the target using Ensembl stable IDs and describe relationships between diseases by mapping them to Experimental Factor Ontology (EFO) terms. The Platform supports workflows starting from a target or disease, and shows the available evidence for target – disease associations. Target and Disease profile pages showing specific information for both target (e.g baseline expression) and disease (e.g. Disease Classification) are also available
Open Molecules Platform
Boehringer Ingelheim has added it's well-characterised non-covalent ATP-competitive inhibitor of glycogen synthase kinase (GSK-3) Bi-5521 to its open molecule platform opnMe.com.
opnMe.com, the new open innovation portal of Boehringer Ingelheim, aims to accelerate research initiatives and enable new disease biology in areas of high unmet medical need by sharing well-characterized, best-in-class, pre-clinical tool compounds.
BI-5521 is a potent and selective ATP-competitive small molecule inhibitor of glycogen synthase kinase 3 (GSK-3), GSK-3β (IC50) 1.1 nM, with demonstrated in vivo activity. Rat pharmacokinetics are available, together with an inactive related compound.
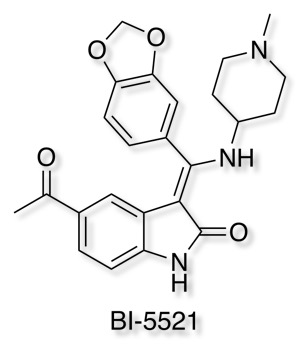
Another useful tool for Target Validation.
How reliable is the literature?
In the past I've mentioned some of the concerns about antibody selectivity, the problems with irreproducible studies and the need for well characterised chemical probes. Elisabeth Bik has been looking at concerns with some of the images in the published literature, The prevalence of inappropriate image duplication in biomedical research publications. mBio 7(3):e00809-16. DOI. her Twitter feed contains yet more examples from the current literature, well worth a browse.
So completing this set, I looked at 101 papers, all published in the same month in the same journal. Of these, 63 contained photographic images (the others had only line graphs and/or tables). Of those, 8 have potential duplications. That is 12.7%
With all the advances in AI and image recognition I'm slightly disappointed that there is not a programme that can do this automatically for Elisabeth.
Neuroscience Chemical Probes
We recently heard that Pfizer is leaving the neuroscience therapeutic area, with a resulting loss of around 300 jobs. This is of course bad news for the scientists involved but I hope the work that was undertaken within Pfizer does not disappear. Chemical probes are critical tools in target identification and validation and arguably even more so in neuroscience. I hope that Pfizer consider releasing some of the well characterised molecules as freely accessible chemical probes, especially if they could also offer a similar but inactive molecule as a negative control. Many of the older tool compounds reported in the literature have been shown to have inadequate selectivity which compromises understanding the biology.
There are many important therapeutic targets within neuroscience but our biological understanding is currently inadequate to justify the investment in drug discovery, a selection of well characterised probes may provide the tools to support the necessary basic biological research.
Updated Target Validation
I've updated the Target Identification/Validation page in the Drug Discovery Resources. In particular I've added a section on the failure to reproduce literature experiments.
Remember this is an absolutely critical step, almost everything else can be fixed.
Target validation and antibodies
A couple of years ago I mentioned an article reviewing antibody selectivity
In 2011, an evaluation of 246 antibodies used in epigenetic studies found that one-quarter failed tests for specificity, meaning that they often bound to more than one target. Four antibodies were perfectly specific — but to the wrong target.
The issue of antibody selectivity has again been flagged as a concern in oestrogen receptor beta research DOI. This is a major target for breast cancer research and there are multiple ongoing clinical trials https://clinicaltrials.gov/ct2/results?term=ERβ.
We here perform a rigorous validation of 13 anti-ERβ antibodies, using well-characterized controls and a panel of validation methods. We conclude that only one antibody, the rarely used monoclonal PPZ0506, specifically targets ERβ in immunohistochemistry.
Applying this antibody for protein expression profiling in 44 normal and 21 malignant human tissues, we detect ERβ protein in testis, ovary, lymphoid cells, granulosa cell tumours, and a subset of malignant melanoma and thyroid cancers. We do not find evidence of expression in normal or cancerous human breast.
Perhaps more worryingly the authors comment.
While our study focuses on ERβ, we do not think that antibodies towards ERβ are significantly poorer than those targeting other proteins, and it is not unlikely that this problem generates similar obstacles in many other fields.
As I wrote on the Target Validation page
Remember this is an absolutely critical step, almost everything else can be fixed.
Target Validation or not
The reproducibility of some target identification/validation studies has been questioned on several occasions and I've flagged up some of the concerns in the Target Validation section of the Drug Discovery Resources. A recent study, reported in Science 28 August 2015: Vol. 349 no. 6251 DOI looking at psychological science, attempted to replicate published work suggests that 39% of effects replicated the original result. Also Amgen, tried to replicate 53 'landmark' cancer studies and failed to replicate the original studies in all but six occasions, Nature 483, 531–533 (29 March 2012) DOI.
A while back a project was initiated to look at reproducibility in cancer, Science Forum: An open investigation of the reproducibility of cancer biology research DOI.
It is widely believed that research that builds upon previously published findings has reproduced the original work. However, it is rare for researchers to perform or publish direct replications of existing results. The Reproducibility Project: Cancer Biology is an open investigation of reproducibility in preclinical cancer biology research. We have identified 50 high impact cancer biology articles published in the period 2010-2012, and plan to replicate a subset of experimental results from each article. A Registered Report detailing the proposed experimental designs and protocols for each subset of experiments will be peer reviewed and published prior to data collection. The results of these experiments will then be published in a Replication Study. The resulting open methodology and dataset will provide evidence about the reproducibility of high-impact results, and an opportunity to identify predictors of reproducibility.
Well some of the early results are in and they make for pretty sobering if not unexpected reading, of the first 7 papers examined, 2 appear to reproduce the original finding to some extent, three show significant differences from the original studies. The results are published in eLife here and there is an editorial here DOI, and they make an important point.
if all the original studies were reproducible, not all of them would be found to be reproducible, just based on chance. The experiments in the Reproducibility Project are typically powered to have an 80% probability of reproducing something that is true.
The key question is of course, is the failure to reproduce these results due to methodological differences not apparent from the described experimental or whether the fundamental result is invalid. At the moment if you are planning to invest in a drug discovery project based on a single publication then Caveat emptor.
47 de-prioritised pharma compounds opens to researchers
A collection of 47 deprioritised pharmaceutical compounds and up to £5 million is being made available to academic researchers through the latest round of the MRC-Industry Asset Sharing Initiative. The collaboration, between the MRC and six global drug companies, is the largest of its kind in the world.
The list of compounds is available here together with brief description of the molecular target and pharmacology, Safety, Tolerability and toxicity information, and any clinical studies that might have been undertaken.
Target Valaidation site update
TargetValidation.org has been updated.
This release brings new web displays and plenty of extra data to assist you in drug discovery and validation:
- 30,591 targets
- 9,425 diseases
- 4.8 million evidence
- 2.4 million target-disease associations
There are also new Web Widgets for both 'RNA baseline expression' and 'Protein Structure' of a target. In the latter, you can now rotate the protein structure, change its colour, zoom in and out, and highlight any amino acid residue:
More information is available on the blog
Chemical Probes Portal Updated
The Chemical Probes Portal has been updated, the new site includes a lot more data about the existing probes, reviewer ratings and their comments.
A chemical probe is simply a reagent—a selective small-molecule modulator of a protein’s function—that allows the user to ask mechanistic and phenotypic questions about its molecular target in cell-based and/or animal studies. These are tools not drugs, they allow scientists to investigate the relationship between a molecular target and the broader biological consequences of modulating that target in cells or organisms. In general the focus is on specificity for the target rather than pharmacokinetics.
Open Targets
A little while back I mentioned the Centre for Therapeutic Target Validation, well it seems that it has now been renamed Open Targets.
The Target Validation platform brings together information on the relationships between potential drug targets and diseases. The core concept is to identify evidence of an association between a target and disease from various data types. A target can be a protein, protein complex or RNA molecule, but we integrate evidence through the gene that codes for the target. In the same way, we describe diseases through a structure of relationships called the Experimental Factor Ontology (EFO) that allows us to bring together evidence across different but related diseases.
There is a video online describing it in more details https://vimeo.com/149309356
This is an absolutely invaluable resource for anyone involved in drug discovery, simply type your query into the text box and submit the query.
This update also bring programmatic access to the data via a series of REST services, the API is fully documented. All the methods are available via a GET request and will serve the output formatted as json. There is a getting started tutorial available.
Target Validation
There has been much discussion about the attrition of drugs in development due to lack of efficacy in man and this in part can be due to poor target validation. That is proof that modulation of the identified target in a model system has the desired impact on biological activity and can be linked to therapeutic utility.
This is an absolutely critical step, almost everything else can be fixed.
For this reason two new resources seem particularly valuable.
The Centre for Therapeutic Target Validation platform (https://www.targetvalidation.org) brings together information on the relationships between potential drug targets and diseases. The core concept is to identify evidence of an association between a target and disease from various data types.
A target can be a protein, protein complex or RNA molecule, but we integrate evidence through the gene that codes for the target. In the same way, we describe diseases through a structure of relationships called the Experimental Factor Ontology (EFO) that allows us to bring together evidence across different but related diseases.The platform supports workflows starting from either a target or disease and presents the evidence for target – disease associations in a number of ways through association and evidence pages.
DisGeNET(http://www.disgenet.org/web/DisGeNET/menu/home) is a discovery platform integrating information on gene-disease associations (GDAs) from several public data sources and the literature doi.
The current version contains (DisGeNET v3.0) contains 429111 associations, between 17181 genes and 14619 diseases, disorders and clinical or abnormal human phenotypes.
Lack of reproducibility with antibodies
A slightly worrying article in Nature, Reproducibility crisis: Blame it on the antibodies.
The lack of reproducibility of published data on potential drug targets has been highlighted on several occasions DOI and it has been suggested that this is a major factor in the failure rate for phase 2 clinical trials DOI.
In almost two-thirds of the projects, there were inconsistencies between published data and in-house data that either considerably prolonged the duration of the target validation process or, in most cases, resulted in termination of the projects.
Antibodies have rapidly become a key tool in understanding and identifying new drug targets and potentially used as biomarkers to identify patients. However it is clear that many of the 2 million commercially available antibodies need to be checked rigorously, with some scientists claiming more than half are unreliable.
In 2011, an evaluation4 of 246 antibodies used in epigenetic studies found that one-quarter failed tests for specificity, meaning that they often bound to more than one target. Four antibodies were perfectly specific — but to the wrong target.
Caveat emptor.
Centre for Therapeutic Target Validation
Target validation is the most critical step in drug discovery because as the chemists will tell you “Most of the other things we can fix”, so I was delighted to hear about the new Centre for Therapeutic Target Validation.
You can read more about it in the Press release
”The Centre for Therapeutic Target Validation is a transformative collaboration to improve the process of discovering new medicines,” says Dr Birney. “The pre-competitive nature of the centre is critical: the collaboration of EMBL-EBI and the Sanger Institute with GSK allows us to make the most of commercial R&D practice, but the data and information will be available to everyone. It is truly exciting to apply so many different areas of expertise, from data integration to genomics, to the challenge of creating better medicines.”
I wish them every success and will be following their work closely.
