Open science ASAP antiviral discovery center
The National Institute of Allergy and Infectious Diseases (NIAID), part of the National Institutes of Health, has awarded approximately $577 million to establish nine Antiviral Drug Discovery (AViDD) Centers for Pathogens of Pandemic Concern. https://www.nih.gov/news-events/news-releases/nih-announces-antiviral-drug-development-awards. One of the centres is the AI-Driven Structure-Enabled Antiviral Platform (ASAP) https://asapdiscovery.org.
ASAP uses artificial intelligence and computational chemistry to accelerate structure-based open science antiviral drug discovery and deliver oral antivirals for pandemics with the goal of global, equitable, and affordable access.
ASAP Overview
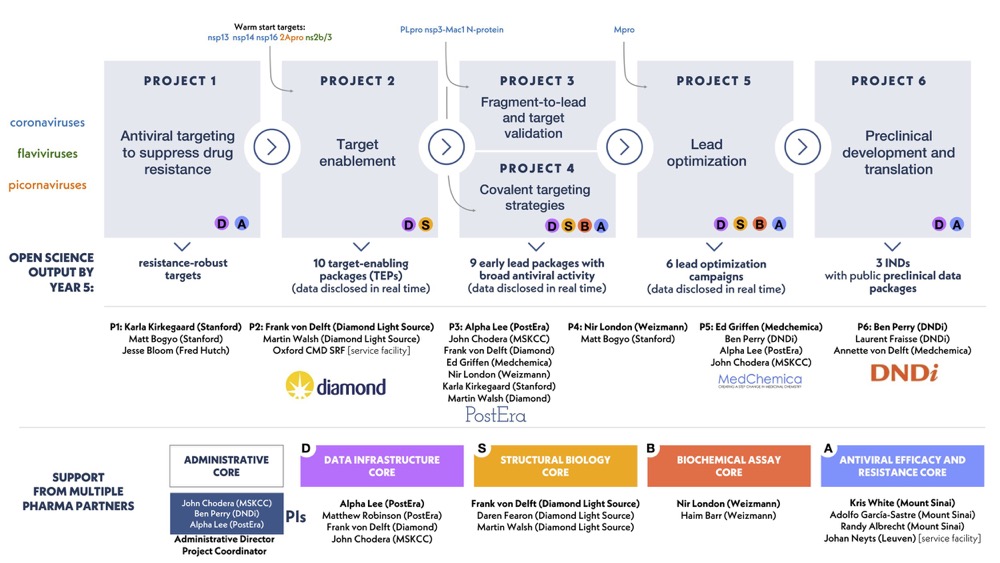
They are currently recruiting for multiple positions, so if you would like to be involved in this exciting programme head over to https://asapdiscovery.org/jobs/.
Open-Source Tools workshops
Registration for the next batch of Open-Source Tools workshops run by the RSC Chemical Information and Computer Applications Group is now open.
https://www.eventbrite.com/e/open-source-tools-for-chemistry-tickets-294585512197?.
These workshops have been enormously popular and the interactions with the instructors have been especially valuable. Details of the next 3 workshops are described below.
All meetings start at 2 pm UK time (5 min break after 1 hour). All run using Zoom Webinar
21 April 2022 PDBe Knowledge Base (David Armstrong)
This workshop explores the Protein Data Bank in Europe Knowledge Base (PDBe-KB https://www.ebi.ac.uk/pdbe/) resource and its tools for the investigation, analysis, and interpretation of biomacromolecular structures. PDBe-KB brings together data from all PDB entries and displays this data as aggregated information for individual proteins, including ligand binding sites, macromolecular interactions and more. Furthermore, this community-led resource brings together structural and functional information from a host of other related resources. In this workshop, you will learn how to use the PDBe-KB aggregated views for proteins to investigate structural and function information for proteins and their associated ligands. We will also demonstrate effective use of novel visualisation components of large-scale structural data on these pages, including 3D visualisation of superposed protein structures with their bound ligands.
19 May 2022 KILFS database (Albert Jelke Kooistra, Andrea Volkamer )
Over the past three decades, six thousand structures of the catalytic kinase domain have been made publicly available via the Protein Data Bank. But to what extent are we making use of this wealth of information? In order to harness this data in a better way and to make it readily available for all to use in their research, KLIFS (https://klifs.net) was constructed. KLIFS, i.e. the Kinase–Ligand Interaction Fingerprints and Structures database, is a structural kinase database that systematically collects and processes all structures of the catalytic kinase domain. With the database, you can - for example - easily get a complete overview of all structures, search for ligands with a specific binding mode, identify analogs or your ligands of interest, collect data for your data mining and machine learning applications.
For this workshop, the developers of KLIFS have teamed up with the Volkamer Lab and therefore the workshop will be divided into two segments. First, Albert J. Kooistra will give an introduction to KLIFS and demonstrate different functionalities of the KLIFS website and the integration of KLIFS in KNIME via the 3D-e-Chem nodes. In the second half, Andrea Volkamer and Dominique Sydow will demonstrate, based on their new kinase-focused TeachOpenCADD workflow, how to assess kinase similarity from different data perspectives. They will emphasize their Python package KiSSim – a KLIFS-based kinase structural similarity fingerprint, and OpenCADD-KLIFS – a Python module to facilitate the integration of KLIFS data into kinase research workflows.
23 June 2022 Scoring of shape and ESP similarity (Ester Heid)
Electrostatic effects along with volume restrictions play a major role in enzyme and receptor recognition. Evaluating electrostatic and shape similarities of pairs of molecules such as proposed versus known ligands can therefore be valuable indicators of prospective binding affinities. This workshop will demonstrate how to compute electrostatic and shape similarities using the open-source tool ESP-Sim (github.com/hesther/espsim, doi.org/10.26434/chemrxiv-2021-sqvv9-v3). Available options for comparing electrostatics will be discussed interactively on selected examples of public datasets, along with advice on embedding and aligning molecules prior to computing similarities.
Open Source Antibiotics
I just thought I'd share this email
Dear Friend of Open Source Antibiotics (OSA),
It's been a busy few months in OSA. Recent activity is being captured in weekly public Zoom meetings (every Friday at 2pm London at https://ucl.zoom.us/j/92800004715), and you can see the details in the recordings of those meetings (like this one) and the associated "Github Issues" (like this one).
But as part of those discussions we were wondering about the best way to update everyone quickly. While OSA uses Twitter, there is no good substitute for a short email. So this is the first, short OSA news email. Three points:
- We have guessed you're interested in receiving occasional emails about OSA. If you're not, just email us back to say you'd like to opt out. Nobody likes spam.
- Please forward this to anyone you think might be interested in antibiotics or drug discovery or open science. As an open project, everything is in the public domain, and everyone is welcome.
- To keep things short, each news email has a limit of three items. If you're interested in learning more, then each project has a wiki (current project status, like this) and the Issue Tracker (current To Do list and discussion, like this).
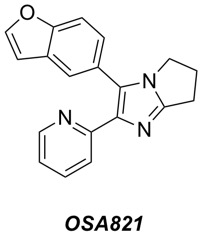
- We have confirmed the activity vs MRSA of the diarylimidazoles (exemplar compound OSA821 shown below), originally discovered and explored by Alvaro Lorente and Bill Zuercher at UNC Chapel Hill. A new potency screen is being performed this week at UCL by Paul Stapleton, and includes about 30 compounds that have been donated to the project via Ben Perry (DNDi). This time our potency assay will include a parallel screen of select compounds vs VRE, to see if there is activity vs other high priority Gram +ves.
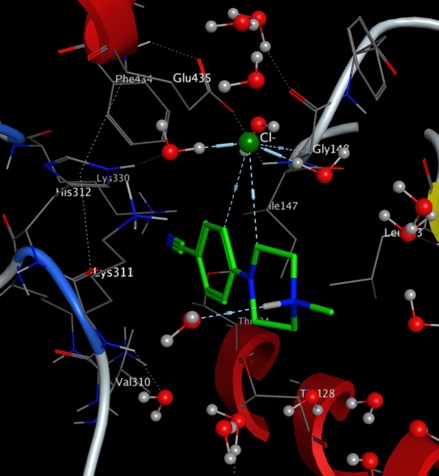
- A key aim of the project is to solve the rapid clearance of the known actives. New data from Sue Charman's lab gave clues as to which compounds to investigate next, and we are finalising negotiations for some pro bono work from a UK company towards identification of possible metabolites.
- The mechanism of action of these compounds is unknown, but Lee Graves's lab at UNC are in the middle of some MIBs experiments that we hope will reveal, by the end of October, some key new insights into how the compounds work.

You can read more about the Open Source Antibiotics on GitHub https://github.com/opensourceantibiotics.
There are currently two projects MurLigase and DiarylImidazoles, everything is in the open and anyone can contribute.
Why not swing by and have a browse.