Do you work with kinases?
If you work with kinases then this free workshop run by RSC CICAG is must for you. There is now a wealth of public domain information about kinases but it is scattered over a multitude of publications and databases. The Kinase–Ligand Interaction Fingerprints and Structures database provides a central repository for all this information. This workshop will guide you through accessing this information.
Register here https://www.eventbrite.com/e/open-source-tools-for-chemistry-tickets-294585512197?.
19 May 2022 KILFS database (Albert Jelke Kooistra, Andrea Volkamer )
Over the past three decades, six thousand structures of the catalytic kinase domain have been made publicly available via the Protein Data Bank. But to what extent are we making use of this wealth of information? In order to harness this data in a better way and to make it readily available for all to use in their research, KLIFS was constructed. KLIFS, i.e. the Kinase–Ligand Interaction Fingerprints and Structures database, is a structural kinase database that systematically collects and processes all structures of the catalytic kinase domain. With the database, you can - for example - easily get a complete overview of all structures, search for ligands with a specific binding mode, identify analogs or your ligands of interest, collect data for your data mining and machine learning applications.
For this workshop, the developers of KLIFS have teamed up with the Volkamer Lab and therefore the workshop will be divided into two segments. First, Albert J. Kooistra will give an introduction to KLIFS and demonstrate different functionalities of the KLIFS website and the integration of KLIFS in KNIME via the 3D-e-Chem nodes. In the second half, Andrea Volkamer and Dominique Sydow will demonstrate, based on their new kinase-focused TeachOpenCADD workflow, how to assess kinase similarity from different data perspectives. They will emphasize their Python package KiSSim – a KLIFS-based kinase structural similarity fingerprint, and OpenCADD-KLIFS – a Python module to facilitate the integration of KLIFS data into kinase research workflows.
These workshops are supported by Liverpool ChiroChem.
KLIFS kinase database
KLIFS is a kinase database that dissects experimental structures of catalytic kinase domains and the way kinase inhibitors interact with them. The KLIFS structural alignment enables the comparison of all structures and ligands to each other. Moreover, the KLIFS residue numbering scheme capturing the catalytic cleft with 85 residues enables the comparison of the interaction patterns of kinase-inhibitors, for example, to identify crucial interactions determining kinase-inhibitor selectivity. DOI.
Statistics
Kinases | 312 |
| Structures (# PDBs) | 5568 |
| Monomers | 12151 |
| Unique ligands | 3497 |
| KLIFS users in Apr-2021 | 897 |
Kinase Inhibitors
I've just updated the page describing Kinase inhibitors, added more on fragments and selectivity.
Kinase Inhibitor Landscape
With over 500 proteins encoded in the human genome it is perhaps not surprising that among enzyme inhibitors, Kinase inhibitors are an increasingly important therapeutic category. The plot below the number of results returned for various string searches of PubMed versus year. Whilst "serine protease inhibitors' (grey) was the highest scoring in 1995 over the intervening years "kinase inhibitors" (red) has risen and is now the highest scoring search string.
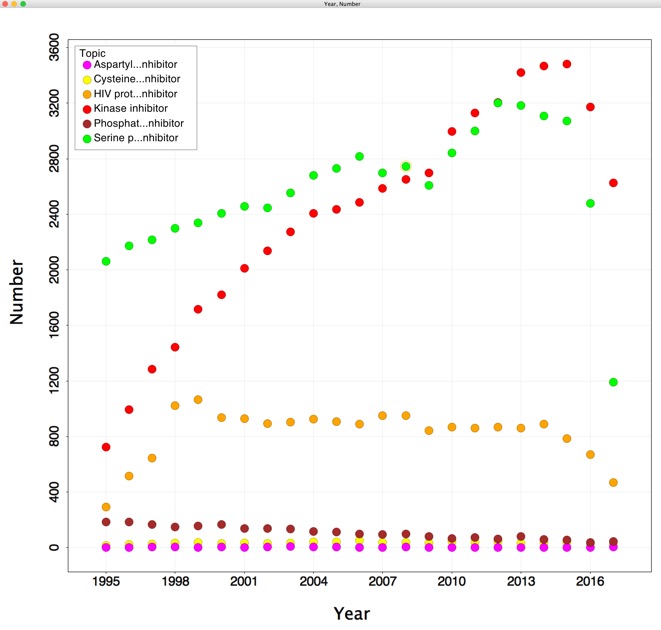
Given that many of the inhibitors target the ATP binding site it is perhaps not surprising that many molecules inhibit multiple kinases, unfortunately this information is not in a readily searchable format. A recent publication "The target landscape of clinical kinase drugs" DOI describes an approach to provide better understanding
To this end, we used a chemical proteomic approach (kinobeads) and quantitative mass spectrometry to characterize the target space of 243 clinical KIs that are approved drugs or have been tested in humans…..The number of targets for a given drug differed substantially. Whereas some compounds showed exquisite selectivity, others targeted more than 100 kinases simultaneously, making it difficult to attribute their biological effects to any particular mode of action.
All drug profiles can be interactively explored in ProteomicsDB and a purpose-built shinyApp.